Enter the matrix
Animal cells must interact with the extracellular matrix in order to divide, manage their shapes, and migrate. Two novel techniques promise to provide more detail on how cells attach to and tug on matrix components.
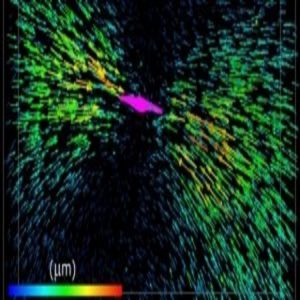
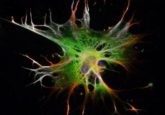
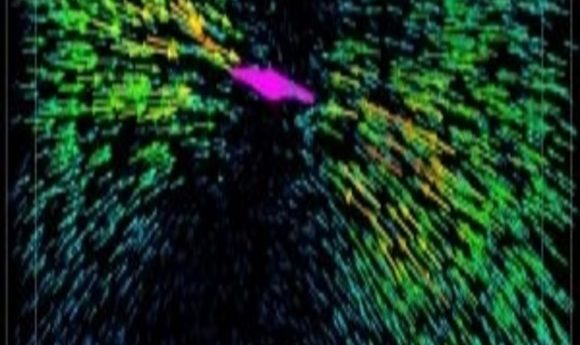
Each arrow represents the displacement of a fluorescent bead tugged by the collagen fibers attached to a breast cancer cell (colored magenta).
Image courtesy of the University of Pennsylvania.

Yue Zhuo uses the photonic crystal enhanced microscope
Image courtesy of G. Pluta/University of Illinois)
Ji Sun Choi, a tissue engineer at the University of Illinois at Urbana-Champaign, studies how blood stem cells differentiate. Unfortunately, scientists don’t have very good markers for the multiple stages of differentiation of these hematopoietic stem cells (HSCs). But Choi, a postdoc in the laboratory of Brendan Harley, uses a new technique, called photonic crystal enhanced microscopy (PCEM), to help her identify those phases of development.
PCEM, described recently in Progress in Quantum Electronics, measures the very bottom 200 nanometers of a cell sitting on a slide (1). Although blood cells are only weakly adherent, they do settle a bit, and the level of adhesion changes as they develop, providing Choi with a real-time biomarker. “With PCEM, you get the information right away,” she said.
PCEM and another recently-published technique using 3-D single-cell traction force microscopy to measure how cells pull on their surroundings (2) offer cell biologists new ways to study cell adhesion. Cells grab onto the extracellular matrix (ECM) using molecules such as integrins, linking their cytoskeletons to the collagen, fibronectin, and other materials in their immediate environment. These interactions contribute to signaling pathways that manage processes such as cell division and growth. They’re also crucial for cell motility. A migrating cell can extend a filopodia to probe the ECM in its target direction and grab hold. Then, other adhesions will have to let go in order for the cell to travel.
Cell types differ in how they interact with the ECM, and as Choi observed, that interaction can change with time. For example, mature blood cells detach from the matrix to enter the circulation.
Alterations to cell–matrix adhesion also occur in disease. For example, stiffening of the extracellular matrix, due to its interactions with cells, underlies liver fibrosis and some tumor types. That’s why a breast tumor manifests as a palpable lump. Cancer cells also rely on cell–matrix interactions to spread around the body.
Crystal-gazing
How do cells grab on, let go, and otherwise interact with the ECM? The inventors of the new techniques hope they can help biologists understand these processes by creating libraries of cell-adhesion interactions for different cell types. At the University of Illinois, bioengineer Brian Cunningham has wanted to use photonic crystals—materials nanostructured with a grating that propagates light along its surface—for imaging since about 2000. His team’s new microscope, which they’ve been developing for about 6 years, is based on photonic crystal sensors that detect the presence or absence of something, such as a cell or virus.
Cunningham and his postdoctoral fellow, Yue Zhao, wanted to get more spatial information out of the sensor, so they married it with a microscope. They coat their slides with a thin layer of photonic crystals and add ECM components such as fibronectin and laminin for the cells to adhere to. When light hits the crystal, it generates an enhanced electromagnetic field that extends approximately 200 nanometers above the slide surface. That’s the “sensing zone,” explained Zhao. If a cell starts to attach to the crystal, it will interrupt that field, generating a signal.
On its own, the microscope would not be able to differentiate the signal of a probing filopodium from a large cell body or a lightly adhered cell from one that’s tightly attached; however, the photonic crystal enhances the contrast between a light or heavy touch on the slide. A spectrophotometer reading the light reflected by the sample will detect a higher wavelength as the cell attaches more tightly.
Zhao admits that she was initially stumped regarding exactly what the PCEM was measuring about the cells in its field of view. After crunching some numbers, she figured out that the light signal indicates, to a good approximation, the “effective mass density” of the cell above. “What we measure is a kind of footprint of the cell,” explained Zhao.
One advantage of this method is that it requires no fluorescent labeling of the cells—as does the similar technique of total internal reflection fluorescence (TIRF) microscopy—so there’s no photobleaching with long-term imaging. Another technique, interference reflection microscopy, images the entire cell body, while PCEM only focuses on that bottom-most surface, noted Cunningham. “We can see how cell attachment points at the cell–ECM interface evolve over long periods of time—hours to days,” he said.
Plus, PCEM is quite sensitive. The researchers tried it out on co-author Choi’s HSCs and could observe the cells’ interactions with the slide surface even though they are weakly adherent at best. “There are actually quite distinctive changes between undifferentiated cells and cells one day after you induce differentiation, versus a week after,” said Choi.
Tug of War
At the University of Pennsylvania in Philadelphia, computational scientist Vivek Shenoy was initially drawn to the question of cell–ECM interactions by the work of a colleague from the medical school, Rebecca Wells, who studies liver fibrosis. After injury, cells deposit ECM and stiffen the tissue to create fibroids. Similarly, as breast cells turn cancerous, they pull on the ECM and stiffen it. “They can actually reorganize the fibers,” said Shenoy.
He and collaborator Mingming Wu of Cornell University wondered how much force the cells exert on the matrix, and vice versa. They applied 3-D single-cell traction force microscopy, a technique that uses fluorescent beads embedded along with cells in a collagen matrix, to measure that force. While a breast cancer cell pulls on the collagen fibers, the researchers image the positions of those beads. Then, the scientists use the drug cytochalasin D to relax the actin cytoskeleton. The pull on the ECM slackens, and the beads move to a location unaffected by the cells. Shenoy and colleagues designed algorithms to compute the force required by the cell to deform the matrix, based on force measurements taken by Wu’s group plus those bead-displacement images.
As the researchers reported in Proceedings of the National Academy of Sciences, they found that the harder the cell pulls, the stiffer the matrix gets. And the stiffer the matrix, the more the cell tugs, creating a positive feedback loop (2).
The result is not entirely a surprise; Clare Waterman of the National Heart, Lung, and Blood Institute called it a “no-brainer” that matrix stiffness and cell pulling would feed back on each other. It was already known that a stiffer matrix promotes more generation of force by cells in a two-dimensional matrix, but the relationship in three dimensions was not so clear-cut, said Shenoy and Wu. Another study reported that the force provided by cells did not depend on ECM stiffness (3).
Building Libraries
“Going forward, it will be exciting for biologists to learn how cell traction force with 3-D biometrics is governed by the cytoskeletal molecules,” said Wu. “This will allow us to gain a handle on controlling cell–ECM cross talk, which will have applications in treating or controlling disease progression.”
Shenoy suspects that more malignant cancer cells may generate the greatest force, and he is currently testing that hypothesis. Perhaps if scientists understood how tumors affect the matrix, they could keep them in check. He suggested that researchers could look at different cell types with this technique to develop a “forceome” of different cell types.
At the University of Illinois, Cunningham and his colleagues have similar plans. “We want to build a quantitative, live-cell footprint library,” said Zhao. Such a resource would help cell biologists better understand the processes of cell adhesion and migration. They could also analyze how different drug treatments affect those mechanisms.
In their paper, Zhao and colleagues reported the very start of such a library, analyzing the adhesion of four cell types. In addition to Choi’s HSCs, they measured footprints for a brain cancer cell line, a mouse epithelial line, and stem cells derived from pig fat tissue. The neurons and epithelial cells adhered tightly and spread rapidly, while the fat stem cells adopted a spindle shape with mid-level adhesion. These data could help researchers identify and classify cell types, the authors suggested.