Hunting for a SARS-CoV-2 biomarker with metabolomics
“Test, test, test” was the guidance offered to combat COVID-19 by Tedros Ghebreyesus, Director-General of the World Health Organization. But before you can test, you must have a marker, a biomarker, that indicates if a person is sick or not.
The SARS-CoV-2 virus, like all biological material, is composed of proteins, lipids, nucleic acids, metabolites and other small molecules. Researchers are using -omics technologies (e.g. proteomics, lipidomics, transcriptomics, genomics) to develop biomarker tests, understand the biology of the novel coronavirus, and provide insight into our immune response to infection. Metabolomics aims to measure all the metabolites and other small molecules in a sample; and it is being used to survey the blood, urine, feces and saliva to identify chemical biomarkers of COVID-19 and better understand the disease.
If you would like to keep up to date with our content on coronavirus, you can sign up for our site here, where you can subscribe to our newsletters for free!
Mass spectrometry is a core technology that is used to perform metabolomics. While you may have never heard of mass spectrometry until reading this article, mass spectrometers are commonly used in clinical diagnostics, so if you have ever had a blood test then you probably have had a mass spectrometry test. Mass spectrometers are ‘scales’ that measure the mass of a chemical to the fourth or fifth decimal place. The power of mass spectrometry-based metabolomics is the ability to provide information about what chemicals are present in a sample and how much is present. However, there is not a mass spectrometry method that can detect all chemicals. As a result, different methods exist to capture different, but complementary, parts of the metabolome.
One common step is to convert the molecules within the sample into gas-phase ions – charged versions of the molecule. There are dozens of ionization methods by which to do this depending on the type of sample (gas, liquid or solid) and chemical properties of the chemicals to be analyzed. Further, chemicals in a complex mixture can be separated from each other over time using another technique called chromatography, which is done prior to ionization. This process enhances the mass spectrometry measurement and aids in identification. In practice, the chemicals in a sample are extracted, purified, chemically modified, etc., prior to performing mass spectrometry. For the purpose of discovery, the most universal pre-analytical steps (extraction, purification, modification and so on) are used in an attempt to capture as much of the chemistry as possible; whereas, for the purpose of diagnosis, very specific pre-analytical steps are used to improve the accuracy and reliability of the measurement.
‘Data rich, information poor’ is how I often describe metabolomics data. The great thing about a mass spectrometer is that it can detect a vast amount of chemical information about a sample, but that ability also quickly leads to an overwhelming amount of data. Often a single study will generate gigabytes of data; however, most of the mass spec ‘peaks’ detected remain unidentified. While the ability to characterize a ‘peak’ has dramatically improved over the last 5 years, in practice, only ~5% of all mass spectrometry signals can be given a chemical name. Researchers have and continue to develop better and better data analysis tools to help combat this shortcoming in information. GNPS has been useful in the discovery of chemicals, with tools such as molecular networking, which link identified and unidentified chemicals together; MASST, which provides researchers with the ability to search public data repositories for additional information (similar to BLAST for sequencing); and others. The development of all these data analysis tools is improved via a strong community of contributors that openly share methods, data and software.
 Refocusing our microbiome lab for SARS-CoV-2 research
Refocusing our microbiome lab for SARS-CoV-2 research
Sarah Allard describes how, in response to the emerging COVID-19 pandemic, they refocused their microbiome lab for SARS-CoV-2 research.
In spite of the challenges of analyzing metabolomics data and the various different ways of performing the mass spectrometry analysis, huge potential exists for understanding COVID-19 infections and finding diagnostic biomarkers. The peer-reviewed literature remains scarce, but new preprints (not peer-reviewed) are emerging; for example, Shen et al. [1] and Wu et al.[2], which present studies with a small number of patient samples from the first country to be affected, China. Shen et al. [1] reported the analysis of serum from 28 healthy, 19 unwell but non-COVID-19, 25 non-severe COVID-19 and 21 severe COVID-19 subjects using metabolomics. Differences in many chemicals were reported including amino acids, carbohydrates, fatty acids and glycerophospholipids. Similarly, Wu et al. (posted on medRxiv [2]) found chemical changes in plasma between 9 fatal, 11 severe and 14 mild cases of COVID-19 as well as 10 healthy volunteers using metabolomics. Given the relatively small number of subjects, the heterogeneous experiences and outcomes of individuals infected with SARS-CoV-2, and the general lack of knowledge about the virus, these reports should be interpreted cautiously; however, it appears that a chemical response to infection is present; specifically, depletion of amino acids in the blood appears to be one target for rapid point-of-care screening tests. Albeit aspecific to COVID-19, such a test may offer a simple, scalable solution for population-scale diagnostic testing.
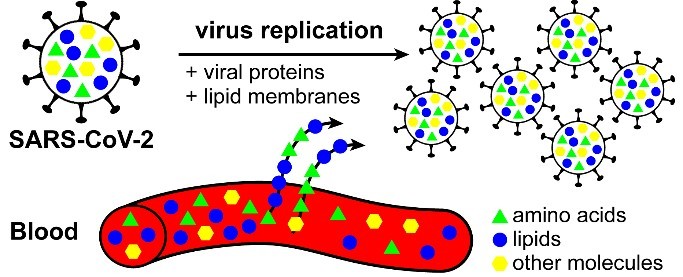
However, there remains a clear need to understand the mechanisms that underpin such signals. For example, why do amino acids and lipids appear to change with infection severity? One could speculate that this stems from the virus’ need for chemical building blocks in order to replicate (Fig 1), but it will take time to understand what the changes mean, as they could also be a result of changes in immune response to the virus, or a combination of both. Aside from biomarkers being used in diagnosis, they can also play an immensely important role in prognosis. Specifically, in Shen et al., [1] the authors stated that the chemical changes were detected days ahead of individuals deteriorating to a critically ill state. Physicians are currently limited in the medications used to fight COVID-19; however, a few days head start may be enough to start medications which interfere with viral replication, such as remdesivir, which is currently being evaluated.
The truth is that we know little and have a lot to discover about COVID-19. More data and validation are required and researchers at the University of California, San Diego (UCSD; CA, USA) are contributing their expertise. The Jain laboratory at UCSD is using metabolomics as a critical tool for understanding what makes people susceptible to COVID-19 infections and how the immune system responds. As previously mentioned, data analysis is difficult and in addressing that the Dorrestein lab at UCSD has launched an effort to compile, and make public, the metabolomics data associated with SARS-CoV-2 to facilitate collaboration with researchers from around the world, including the COVID-19 Mass Spectrometry Coalition with more than 220 scientists from 18 countries. There is immense potential for metabolomics in diagnostic and prognostic testing of COVID-19 that are complementary to the ongoing global efforts to develop nucleic acid and antigen testing.