Using computer models to outsmart spike proteins in SARS-CoV-2
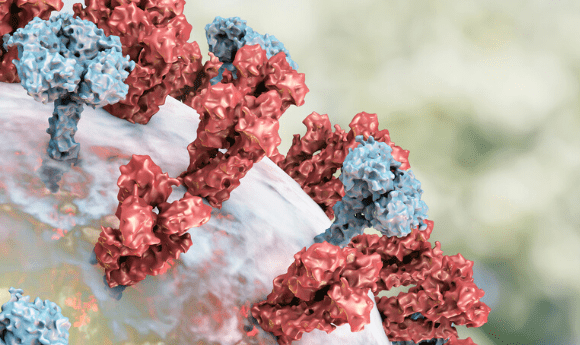
Researchers from the University of Illinois (USA) have developed a computer model to test whether their peptide inhibitor treatment would be effective at blocking the spike protein found on SARS-CoV-2.
Found on the surface of the virus, the spike proteins of SARS-CoV-2 bind to the ACE2 receptors of human hosts, allowing the virus to gain entry to and therefore infect the cell. Peptide inhibitors offer a potential mechanism to prevent infection, by mimicking the protein binding domain of the ACE2 receptors and binding to the spike proteins instead.
To model this mechanism and determine whether it worked in practice, researchers developed a computer model that could simulate the binding actions and identify the most promising inhibitor compound.
Visit our sister site The Nanomed zone to find out more
 Predicting the course of a pandemic: when will COVID-19 end?
Predicting the course of a pandemic: when will COVID-19 end?
Data scientists have utilized artificial intelligence to create data-driven predictions of the trajectories of COVID-19 in different countries, ultimately predicting when COVID-19 will end.