Expanding our insights into the planktonic universe
Original story from the European Molecular Biology Laboratory (Heidelberg, Germany).
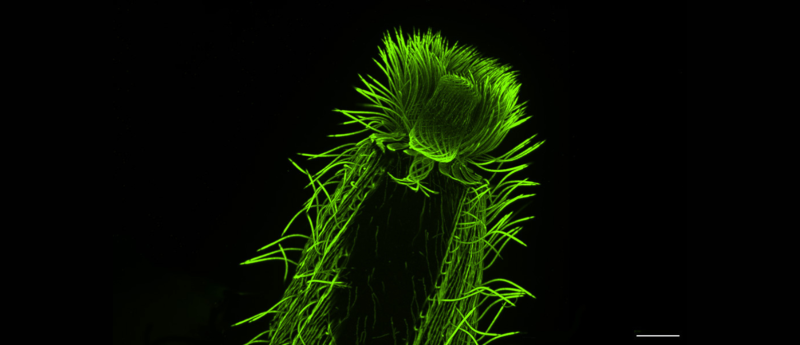
A new study provides unprecedented insights into the cellular architecture of over 200 species of plankton using ultrastructure expansion microscopy.
One day in 2020, in the middle of the COVID-19 pandemic, EMBL (Heidelberg, Germany) Group Leader Gautam Dey received a Zoom call from his collaborator and fellow cell biologist Omaya Dudin. Dudin, who led a research group at École Polytechnique Fédérale de Lausanne (EPFL; Switzerland) at the time, had just succeeded in optimizing a relatively new technique to observe the cellular architecture of a marine microorganism.
Dudin had been working with this particular group of marine microbes – Ichthyosporea – for over six years. However, visualizing its inner structures had been a challenge. Unlike for more commonly studied organisms, there were no genetic tools to make certain structures light up under the microscope. Nor did antibody-staining, the other main technique in the biologist’s toolkit, work well in this organism, as its cell wall prevented antibodies from penetrating the cell.
However, the new technique called expansion microscopy – first developed 10 years ago by scientists at MIT (MA, USA) and further optimized into ultrastructure expansion microscopy (U-ExM) for exploring sub-cellular ultrastructure by scientists at the University of Geneva (Switzerland) – had somehow made this cell wall permeable, letting antibodies enter, and the organism’s inner structures could now be clearly visualized and studied. Expansion microscopy works by physically ‘expanding’ biological samples. These samples – which can contain single-celled organisms, cells or tissues – are first embedded in a clear gel. Then, the gel is allowed to expand by absorbing water. Many of the cell’s internal structures remain intact during the process and expand more or less proportionally, allowing scientists to ‘magnify’ the sample four or even 16 times without needing to resort to lenses.
A collaboration to apply this technique to all microbial eukaryotes was born and 3 years later, the pair along with colleagues have generated an almost encyclopedic knowledge of hundreds of species and are on their way to creating a planetary atlas of plankton.
 The risks of methylmercury contamination in marine ecosystems
The risks of methylmercury contamination in marine ecosystems
Climate change may be increasing the spread of a neurotoxin, methylmercury, in the ocean.
Exploring the planktonic universe
Dey and Dudin are particularly interested in investigating a certain group of microbial eukaryotes: plankton. These tiny organisms, found in the Earth’s oceans, form an irreplaceable part of the oceanic food chain and generate most of the planet’s oxygen. Their diversity already spans tens of thousands of species, and that’s only among those described so far, with many more waiting to be discovered.
Until recently, genomic sequencing was the strongest tool in researchers’ arsenal to study planktonic diversity. Available imaging methods were either high-resolution or high-throughput, but rarely both. Many of these organisms could also not be cultured under laboratory conditions and had no genetic tools available.
“Thus, the fields of cell biology and protistology diverged over the last 40 or so years, with one focusing on increasingly high-resolution microscopy methods, while the other specialized in genomics,” Dudin explained.
For the collaborators, the first opportunity to bridge this gap came during the EMBL-led Traversing European Coastlines (TREC) expedition. One of the expedition’s first stops was in Roscoff (France), where the Station Biologique hosts one of the most complete culture collections of marine microorganisms in Europe. The team asked Ian Probert, the facility’s manager, how many samples they might receive to run an expansion microscopy pilot, expecting about 20 species. Instead, the facility offered them more than 200 species ready to be processed for expansion microscopy.
“We spent three days and nights just fixing those samples. This was a treasure trove we could not let go of,” commented Felix Mikus, who completed his PhD from the Dey Group and is now a postdoc in Dudin’s current lab at the University of Geneva.
Using these samples, as well as a second culture collection from Bilbao (Spain), the researchers went on to perform one of the most extensive investigations to date of the diversity of the cytoskeleton – the filamentous network that forms the underlying structure of eukaryotic cells. In particular, the researchers looked at microtubules – hollow, tubular filaments that help the cell not only maintain its structure but also divide and move. They also studied centrins, a family of proteins found in the structures that organize microtubules inside cells.
This study was the first step in PlanExM, a TREC plug-in project that aims to reveal the world of planktonic ultrastructural diversity with expansion microscopy.
According to the researchers, this analysis not only helps us understand the fundamental principles that underly the organization of a eukaryotic cell, it also offers clues to the evolutionary history of the cytoskeleton architecture in these species. Moreover, it demonstrates the potential of expansion microscopy as a powerful tool to study complex samples, including those collected directly from marine ecosystems.
“U-ExM is transforming how we explore protist ultrastructure,” shared Armando Rubio Ramos, co-first author of the study and postdoc at the University of Geneva. “By combining this technique with high-throughput imaging and comparative analyses, we can begin to decode how cellular architecture has diversified across evolution. It’s a bridge between molecular data and the physical organization of life at the microscopic scale.”
“The next step is to selectively look deeper into certain species within this broad collection to answer specific questions, such as understanding how mitosis and multicellularity evolved and the phenotypic diversity that underlie major evolutionary transitions,” Dudin concluded.
This article has been republished from the following materials. Material may have been edited for length and house style. For further information, please contact the cited source. Our press release publishing policy can be accessed here.