Structural blueprints for bacteria
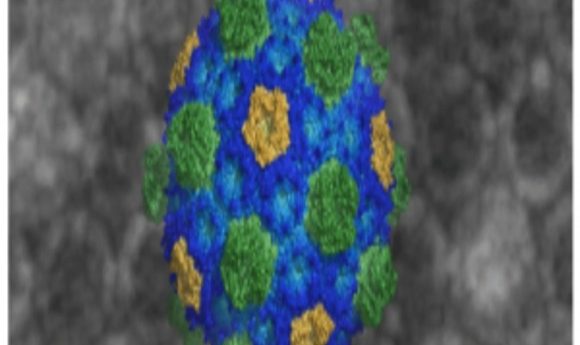
For the first time, researchers captured atomic-level images of organelle microcompartments in bacteria, providing the clearest picture yet of how they are assembled.
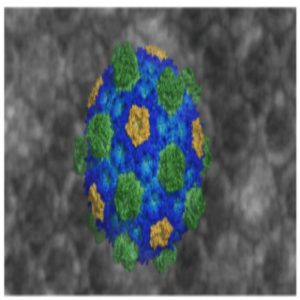
A representation of the bacterial microcompartment membrane showing structures of its constituent proteins.
Credit: Markus Sutter and Cheryl Kerfeld, Michigan State University and Berkeley National Laboratory
Bacterial microcompartments (BMCs) control a range of biochemical activities within bacterial cells, from CO2 fixation to arresting toxic metabolites that may threaten cell health. Until now, researchers have lacked the structural blueprints to understand how BMCs assemble and function in detail.
In a new study published in the journal Science, researchers unveiled the first crystallized BMC shell structure, outlining its spherical membrane-like bioarchitecture, which is made up exclusively of proteins, and revealing assembly rules at atomic resolution. The authors suggest that understanding the detailed shell’s anatomy may help bioengineers build synthetic organelles, fight pathogenic bacteria, or improve the performance of “good” bacteria and drug therapies in the future.
“If you imagine these pieces as Lego building blocks, we had previously known what the individual blocks making up these protein shells looked like in isolation, but had never seen them snapped together,” said Cheryl Kerfeld, who led the study at Michigan State University and Berkeley National Lab. “Understanding what this protein-based membrane looks like has given us the rules for assembling these microcompartment membranes, and this could enable us to engineer compartments for applications like drug delivery.”
The team spent over 2 years obtaining high-quality crystals of an intact shell using proteins from the oceanic bacteria strain Haliangium ochraceum. With the X-ray diffraction data, the team identified how five different types of proteins are arranged to form the micrcompartment membrane. Ultimately, the proteins formed a 20-sided icosahedral shell, which is large enough to contain 300 average-sized proteins.
“Other than viral capsids, this is the largest protein assembly ever crystallized,” said Kerfeld. “We now have a much better idea of how these hexamers, pentamers, and pseudohexamers fit together, and we may now use that to control assembly or even disassembly of these shells.”
Unlike lipid-based organelles in eukaryotic cells, BMC membranes are constructed entirely of a selectively permeable protein layer that can cordon off certain toxic metabolites or protect sensitive enzymes from the rest of the cell. These shells can enhance CO2 fixation in photosynthetic cyanobacteria by encapsulating the oxygen-sensitive enzyme Rubisco. However, because some bacteria make use of BMCs to break down carbon compounds for energy, the organelles can also provide fuel for pathogenic bacteria.
“A lot of pathogenic bacteria make use of bacterial microcompartment-based metabolism to outcompete good bacteria in an environment where they are able to break down something that the good bacteria cannot consume,” explained Kerfeld. “We want to find compounds that interrupt that shell assembly or permeability, which could be used as a new type of antibiotic. Now that we know how the proteins in these shells fit together, it provides a starting point for disrupting the shells in pathogens.”