Rapid screening of viral inhibitors could improve response to outbreaks
An efficient strategy enables the identification of potent inhibitors that target critical viral proteins called deubiquitinating enzymes.
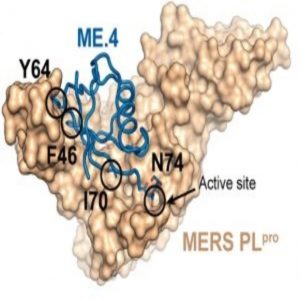
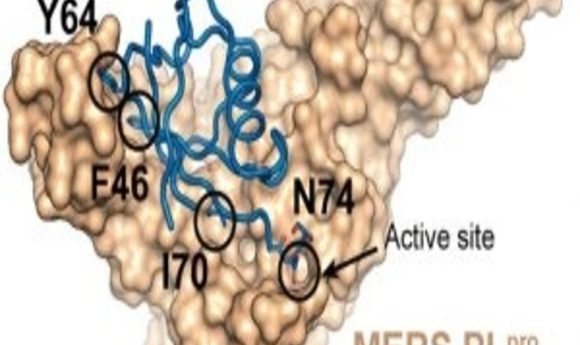
Structural basis for UbV inhibition of MERS-CoV PLpro (1).
Recent outbreaks of viruses such as Ebola and Zika have shown that the global health community remains unprepared to quickly respond to emerging infectious diseases. In a study published in PLOS Pathogens, researchers addressed this problem by developing a method to rapidly identify potent inhibitors of viral proteins called deubiquitinating enzymes, which play a critical role in promoting viral replication and suppressing host immune responses.
“This study is important because it shows for the first time a rapid and efficient platform to target viral proteins for inhibitors that can be used as antiviral agents,” said first author Wei Zhang, a postdoctoral fellow in Sachdev Sidhu’s lab at the University of Toronto. “While we develop vaccines for healthy people to prevent viral diseases from happening, we now have a method to quickly develop drug-like molecules to treat infected patients.”
Ubiquitin is a protein that plays a critical role in a wide variety of cellular processes. As such, it is not surprising that viruses have evolved ways to disrupt the ubiquitin system to promote infection and replication in host cells. A broad range of viral pathogens produce deubiquitinating enzymes, which remove ubiquitin from host proteins and thereby suppress host immune responses. Although viral deubiquitinating enzymes are ideal drug targets, their structural similarity to human deubiquitinating enzymes has hindered the development of safe, highly selective, small-molecule inhibitors suitable for clinical use.
Zhang and his collaborators developed a strategy to rapidly identify potent protein-based inhibitors that specifically target viral deubiquitinating enzymes. Specifically, the researchers used a phage-displayed library of billions of ubiquitin variants to efficiently screen for inhibitors that bind with high affinity and specificity to deubiquitinating enzymes produced by the Middle East respiratory syndrome coronavirus (MERS-CoV) and the Crimean-Congo hemorrhagic fever virus.
“We used protein engineering to turn ubiquitin into a completely different protein, termed ubiquitin variant, which binds viral proteins with super high affinity to inhibit the catalytic activity of those proteins,” Zhang explained. “And we used phage display to enable quick and efficient screening of functional molecules.”
Notably, ubiquitin variants generated against MERS-CoV almost completely abolished the replication of this virus in host cells. Meanwhile, none of the ubiquitin variants generated in this study likely cross-reacted with human deubiquitinating enzymes, demonstrating their potential therapeutic safety.
As noted by the authors, phage display yields potent and selective viral inhibitors in weeks, whereas small-molecule approaches can take years to implement and often fail. This key advantage could allow therapeutic development to keep pace with the continued emergence of pathogenic viruses and limit their pandemic potential. “Our approach could be employed potentially for any viral intracellular proteins and quickly adapted to any emerging or re-emerging viruses,” Zhang said.
But the implications for human patients will likely not be evident for a long time. One major therapeutic hurdle is the lack of efficient ways to deliver proteins inside cells. Although Zhang and his collaborators are working on this problem, ubiquitin variants are best viewed as tool compounds that can enable drug discovery rather than bona fide drug candidates—at least for now.
“Clearly, this can be used to develop antiviral therapeutics for humans, but in the short term, we will focus on viral diseases of plants and livestock because of the transgenic technology to easily deliver the ubiquitin variant inhibitors,” Zhang said.