Untapped peptide potential
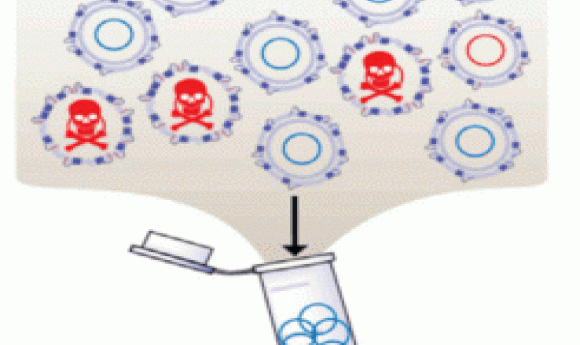
A new high-throughput screening platform rapidly tests and identifies antimicrobial peptides that act against multi-drug-resistant gram-negative bacteria.
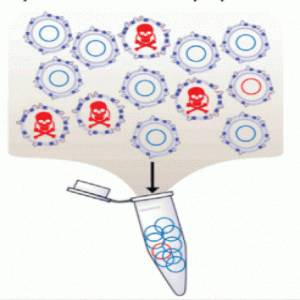
Bacteria expressing peptides with antimicrobial activity drop out of the population (1).
The antibiotic arms race is in full swing with the expected rise in antibiotic-resistant bacteria. It’s been nearly 40 years since the last discovery of a new class of antibiotics, and the clock is ticking. Antimicrobial peptides are integral to the innate immune response and are beginning to resurface as promising new therapeutics. Although current platforms can effectively screen for a peptide’s ability to bind to a target cell, available tools for screening peptides for new leads in a biologically relevant manner are lacking.
Now, researchers from The University of Texas at Austin have published a paper in the journal Cell showcasing a new method for the rapid, high-throughput screening of antimicrobial peptide functionality. Using this method, the researchers identified a new class of lead peptides with distinctive mechanisms, no longer confining researchers to the study of naturally occurring peptides.
In the early stages of treatment, antibiotics bind to the cell membrane of the bacterium. The researchers wanted to emulate this scenario in their own drug screening platform. “The challenge was that we wanted to actually present the peptide to the bacteria in a therapeutically meaningful way,” said Bryan Davies, lead author of the paper.
To do this, the researchers developed a screening platform called Surface Localized Antimicrobial Display (SLAY) that first delivers peptides to a gram-negative bacterial surface. “The idea was to have the bacteria synthesize and self-test the peptide on itself,” said Davies. By commandeering bacteria’s natural propensity to synthesize peptides, the researchers were able to cut down on costs and time. “And then the system presents [the peptide] on the cell surface of the bacteria in a tetherball fashion where the peptide can then come back and interact with the bacteria that made it,” said Davies. Here, the peptide will either elicit an antimicrobial effect and kill the bacterium or not, which is measured using next-generation sequencing.
Using SLAY, the researchers screened a library of 800,000 peptides found that 1.7% showed antimicrobial activity–that’s over 8000 untapped peptides! Further synthesis and testing showed that a majority of these lead peptides are fully synthetic with a physiochemical profile beyond what is found in nature. “We’ve identified peptides that have very new sequences and chemistry [compared to] what were used to,” said Davies. One day soon, researchers may add synthetic antimicrobial peptides to their arsenal for fighting antibiotic resistance.