Fluorescent imaging system sheds light on cellular autophagy
New technique allows real-time imaging of ubiquitination by gene tagging with split-fluorescent proteins.
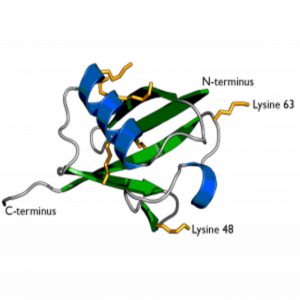
The secondary structure of ubiquitin showing the two previously known residues Lysine 63 and Lysine 48
Researchers from the Tokyo Medical and Dental University (Tokyo, Japan) have published a study in Autophagy revealing a new technique to live image ubiquitin tail formation by genetically engineering ubiquitin, which induces fluorescence during ubiquitination.
This process can help characterize the residues used for linkage by ubiquitin. The residues determine how the protein will degrade whilst undergoing autophagy.
Homeostasis is the system by which cells maintain balance through physiological processes. One of these processes is autophagy whereby toxic or unnecessary material, including proteins, is degraded.
Ubiquitin is a signaling protein that binds to the surface of such material, targeting it for degradation. Ubiquitin molecules attach to each other to form a polyubiquitin chain of repeating units. This process, termed ubiquitination, signals that the protein is suitable for autophagy. However, there are eight different potential residues used in ubiquitination and these are important in determining the functionality of autophagy.
Author Yoichi Nibe commented, “We know that the particular residue used for linkage can determine which degradation pathway a protein will follow during autophagy, but so far only two of these residues have been characterized. The function of the other six is largely unexplored territory. What we needed was a system that would allow us to analyze any ubiquitin residue we choose.”
In order to determine which residues were forming the polyubiquitin chain, the researchers genetically engineered the ubiquitin molecules by attaching a split protein, Kusabira Green. The two halves of the split protein fluoresce when they interact. Hence, when two corresponding residues are formed in the polyubiquitin chain during ubiquitination, they emit light that can be captured and recorded live.
Corresponding author Shigeru Oshima explained, “This system allows us to visualize ubiquitin assembly in real time, in living cells. We can tightly control the expression of ubiquitin using experimental induction, which is ideal for studying a highly dynamic process such as ubiquitination. Most importantly, though, we can manipulate individual residues of ubiquitin to force specific chains to form in the cell.”
Oshima is very hopeful about the potential of this technique, as it has already identified a previously unknown residue, lysine 33.
“The system is very versatile. Given the complexity of ubiquitination, we expect to uncover some interesting functions in the future.”