No two shells are the same
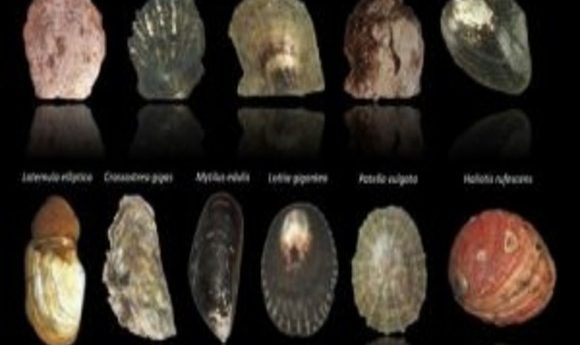
Beachcombers know that seashells appear in an endless array of colors and shapes, and researchers have just discovered that this variation extends to the molecular level. In fact, each shell is as unique as a fingerprint.
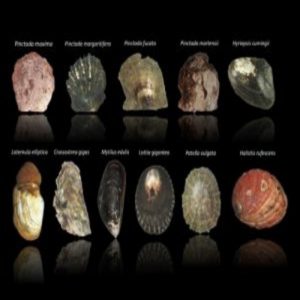
Transcriptomes were generated for these 11 mollusks – the largest genetic study of shelled organisms ever performed.
Credit: Felipe Aguilera
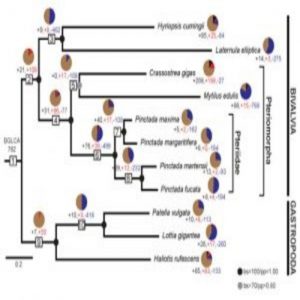
Evolutionary dynamics of mantle gene families – the mantle secretome – over mollusk evolution. BGLCA is the common ancestor of bivalves and gastropods surveyed in this study (1).
Every morning on his way to class, Felipe Aguilera, a budding marine biologist at the University of Valparaiso on the coast of Chile, took a stroll across the beach. While shuffling down the rocky shoreline, he searched for tiny logarithmic spirals and other complex structures that once harbored vulnerable critters. His shell collection grew, alongside a newfound fascination with the phylum Mollusca.
Aguilera’s growing intrigue with seashell fabrication, patterning, color, and diversity led him to other coastal schools around the world. In 2010 he joined a small group of shell experts to embark on a great enterprise to decipher the molecular blueprint behind shell architecture. To accomplish this, the group performed one of the largest molluscan biomineralization surveys ever, collecting 11 transcriptomes of bivalves and gastropods, and culling new insights that challenged long-held hypotheses on shell formation.
“Many [experts] suggested the presence of a deeply conserved biomineralization ‘toolkit,’ so we expected to find it,” said Aguilera, lead author of a new study in the journal Molecular Biology and Evolution (1), who is now a postdoctoral fellow at the Sars International Centre for Marine Molecular Biology at the University of Bergen in Norway. “Surprisingly, we found no unique model or common molecular toolkit behind the making of every shell. It doesn’t matter the level of relatedness among shelled mollusks, each species seems to evolve its own unique genetic instructions to fabricate the shell.”
These genetic instructions are particularly special because each mollusk shell has different combinations of “ancient” and “novel” genes. Like a fingerprint, mollusk shells have a distinct identity.
A Repertoire of Genes
Mollusks first evolved in the ocean about 500 million years ago before making their way to surface ecosystems. There is a multitude of extant mollusk species – over 130,000 and counting – making this group the second largest phylum after arthropods (2). The name was derived from the Latin word mollis meaning “soft” and was coined by Aristotle to describe cuttlefish and octopuses (3). Both of these animals are squishy and flexible, but they also have protective, internal shells. Not all mollusks bear a shell, but the ones that do are called “conchiferans”—consider the iconic “conch” shell.
Mollusks, in general, sparked wonder in marine biologists because of their tremendous diversity, but conchiferans are especially interesting to both scientists and shell collectors for their beautiful, complex and sturdy armors.
Aguilera’s graduate school adviser, Bernard Degnan of the School of Biological Sciences at the University of Queensland in Australia, worked for over a decade to understand shell biomineralization by looking at genes expressed in a mollusk’s mantle—the organic outer layer involved in creating the shell. Conchiferan shells consist of 95 to 99 percent calcium carbonate minerals that the animal obtains from seawater; the rest is organic material secreted by the mantle (4).
During graduate school, Aguilera gathered the transcriptomes of eight adult bivalves (clam- and oyster-like sea animals) and three mature, shelled gastropods (sea snails). With the 11 mollusks, Aguilera and his lab mates traced the evolutionary history of the mantle secretome in search of the aforementioned molecular “toolkit.”
By using a phylostratigraphic approach – a method that helped the researchers investigate genes starting from the origin of cells all the way to the 11 modern species– the team discovered that the mollusks develop their shells using a repertoire of co-opted ancient genes from before the evolution of conchiferans and a suite of rapidly evolving novel genes specific to each modern species. Moreover, the team noticed that for both bivalves and gastropods, over 80 percent of the gene families were co-opted ancient genes, but the remaining novel or species-specific genes were the main drivers of mantle secretome evolution (1).
“Some of these species have diverged from each other on a certain timescale similar to the way we’ve diverged from chimps and gorillas,” Degnan said. “The animals we studied share a distant common ancestor, so they’re related, but the genes behind each animal’s mantle secretome holds different instructions for each shell. We sort of debunked a leading hypothesis.”
For example, the team analyzed transcriptomes of four pearl oysters that diverged within the last 20 million years—some relatively young Pinctada species. The oysters’ novel genes comprised a huge portion of their secretomes which caused a rapid change to each shell’s look and feel.
“I also showed that many lineage- or species-specific genes are subjected to positive selection which supports the fast-evolving nature of the mantle secretome,” Aguilera said. This idea suggests that not only does the mixing of old and new gene families make for neat shell aesthetics, it provides an evolutionary advantage for each species’ survival (5).
The Key to Diversity
So what exactly do novel mantle secretome genes encode to make so many beautiful and durable shells? Secreted proteins from each species’ mantle secretome contains repetitive low complexity domains (RLCDs), which are prone to insertion and deletion errors during genome replication, thus allowing each animal to form a completely unique secretome and shell.
RLCDs are actually quite common in a wide range of organisms, particularly if they are rich in glycine, showing up in biomineral structures such as eggshells, cuticles, spider silk, and even cell walls (6). In general, proteins containing RLCDs have several specific biomineral functions: they can create both rigid or flexible protein scaffolds for different types of support and bind to calcium ions when protein environments are acidic (4).
Conchiferans have their own sets of RLCD-containing proteins that are encoded by either completely novel gene families or some combination of novel and ancient gene families. The researchers believe the high prevalence of RLCDs comprising these secretomes is key to explaining molluscan evolution (1).
“It’s likely that the rapid diversification of these proteins contributes to the diversity and patterning of mollusk shells, as well as the diverse range of mechanical and physical properties of shells,” Aguilera explained. “The mantle secretome is evolvable, allowing for variation that underpins the spectacular diversity of molluscan shells.”
Branching out on the Family Tree
Though the survey was extensive and took about 2 to 3 years to analyze, the story of mollusks remains incomplete. Aguilera and Degnan are interested in other areas like witnessing the changes of the gene regulatory network (GRN) during mollusk life stages, such as larvae. The 11 mollusks in question were completely mature animals, so experts don’t know whether the same genes are expressed throughout life and if RLCDs play a role prior to the shell-making stage of life. According to Aguilera, no previous studies have looked into GRNs of different life stages, so the field is missing some crucial information.
Additionally, the team is interested in investigating the genomes of the other half of Mollusca—the aculiferans, which have shell plates or spicules instead of large shells. Since the shelly creatures Aguilera’s team has already studied have numerous co-opted ancient genes older than the first conchiferan to evolve, what genetic patterns might they share with aculiferans? Perhaps these more exposed mollusks have more expression of ancient genes than their shelled counterparts.
“These comparative studies are becoming larger and give us so much more detail about the evolutionary processes of biodiversity,” Degnan said. “In order to reveal further secrets of the mantle secretome, we’re changing the scope of how we think about biodiversity; we consider less about ‘survival of the fittest’ and more about ‘arrival of the fittest.’”
Researchers have some ideas about how and why rapid evolution takes place, but knowing when changes occur in an organism’s evolutionary history explains a whole lot more. These insights become especially important in a modern world dealing with ocean acidification and knowing when a next generation of mollusks can come forth and deal with new challenges