Cas9 in DNA-cutting action
New research shows that Cas9 cuts DNA differently than scientists believed. How will a better understanding of the enzyme improve CRISPR-Cas9 genome editing technology?
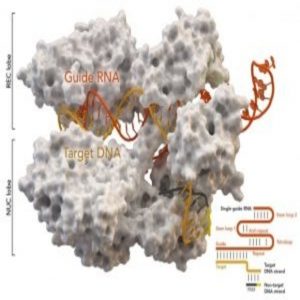
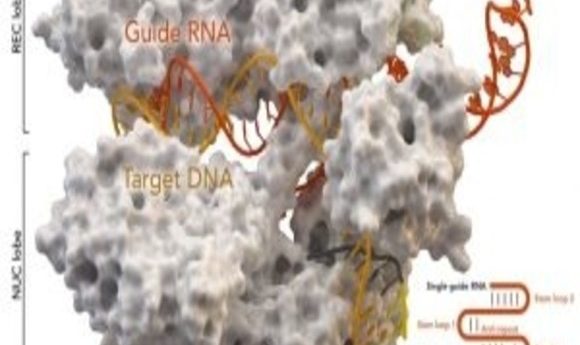
University of North Texas researchers use Maverick to produce the first-ever all-atom computational simulation of Cas9 activity from Streptococcus pyogenes (spCas9), complexed with a single-guide RNA (sgRNA), on a double strand DNA (dsDNA) substrate. Credit: Jin Liu and & University of North Texas.
Although researchers have thoroughly explored and developed many potential applications for CRISPR-Cas9 genome editing technology since its introduction in 2011, how the Cas9 protein behaves and cuts DNA at a molecular level hasn’t been clear.
Now University of North Texas researchers provide new details about the precise location at which the enzyme cleaves DNA and evidence of staggered-end DNA breaks left after cutting. The team published their findings in Scientific Reports.
“This research is the first to capture the active state of the Cas9 enzyme,” said Jin Liu, who led the study. “We uncovered the critical role of magnesium ions in the CRISPR-Cas9 system, and also showed that the active site where Cas9 cuts DNA is not where many people assumed.”
To observe the protein in action, the team turned to the Maverick supercomputer at the Texas Advanced Computing Center (TACC), which is capable of creating molecular dynamics simulations of Cas9 activity at femtosecond resolution based on analysis of a newly crystallized Cas9-sgRNA-dsDNA structure obtained from the Protein Data Bank.
To activate Cas9, Liu’s team introduced magnesium (Mg2+) ions at certain genomic sites they believed would bridge the gap between the active site of Cas9 and the DNA, effectively instigating cleavage. For each placement configuration of the Mg2+ ions, the team produced at least 6 parallel simulations to ensure that they caught when and where the protein became active.
“We initially put the ions at the 3rd nucleotide position, which many believed would cause the Cas9 protein to become active,” explained Liu. “However, to our surprise, conformational changes only happened once we placed the ions at the 4th nucleotide position, leaving the enzyme and DNA in a perfect position for the cleavage.”
“This is significant because if Cas9 cuts the non-target DNA strand at the 3rd nucleotide it would produce blunt-ended breaks, but at the 4th it generates staggered-ended breaks that are more controllable for genome editing,” Liu added.
For now, Liu’s team has only simulated Cas9 interaction with partial DNA at non-target sites of DNA. The team now looks to use Maverick to simulate interactions of Cas9 with whole DNA at both non-target and target sites.
“This could be critical for helping us shed light on new ways to improve Cas9’s specificity, efficiency and help us cut DNA in a more controllable way,” said Liu.