Solving the limitations of ‘bottom-up’ proteomics

The PRISM laboratory research team at Lille University (France), describe their novel ‘top-down’ proteomics approach to analyze and localize intact proteins.
The PRISM laboratory group: (from left to right) Dr Jusal Quanico, Dr Julien Franck, Professor Michel Salzet, Dr Maxence Wisztorski, Professor Isabelle Fournier, Antonella Raffo-Romero.
The PRISM Laboratory (the name represents proteomics, inflammatory response, and mass spectrometry), is a unique worldwide laboratory creating and developing novel technologies, new fundamental biological concepts, new diagnostic tools and novel treatments, all tuned for personalized medicine in oncology and neurology. It is structured around two main pillars; one devoted to technological innovations and the second to therapeutic innovations. PRISM is associated to the Department of Surgery and Cancer at Imperial College London (UK).
Modern protein analysis, called proteomics, allows large-scale identification and quantification of proteins in complex mixtures extracted from cells, tissues and biological fluids. The most common technique is called ‘bottom-up’ proteomics, involving the use of enzymes digesting proteins into small pieces that are ultimately analyzed and sequenced using tandem mass spectrometry combined with bioinformatics tools. However, this approach has major drawbacks.
First, the information about the intact size of proteins is lost because of the digestion process. Second, the potential modifications of proteins can be hard to detect and often require specific sample preparation and analysis. Third, the protein localization within a tissue is lost because tissues are usually processed as a whole and thus information about the tissue heterogeneity and complexity are discarded. Fourth, proteins are identified using closed databases that limit the discovery of novel proteins. These key aspects are even more crucial in the context of clinical tissue sample analysis, where protein expression, localization, form and modification can be critical to understanding molecular pathways involved in pathogenesis and in defining new biomarkers for diagnostics and prognosis.
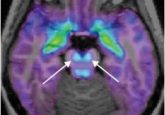
In this context, we developed an approach to analyze and localize intact proteins from precise areas of biological tissues using limited amounts of sample by ‘top-down’ proteomics. First, the tissue sample is analyzed using matrix-assisted laser desorption ionization mass spectrometry imaging (MALDI-MSI), which is a powerful molecular histology tool employed to distinguish regions within a tissue (e.g., tumorous and benign regions).
Once regions are defined, proteins are then extracted in micro-volumes and analyzed using nano-liquid chromatography coupled to a mass spectrometer. This leads to the identification of hundreds of intact proteins, which can be further studied for their biological functions and molecular pathways. By this approach, we can expose proteins that harbor modifications with varying abundances within regions, which implies specific biological involvements and activities.
We are also able to identify many truncated forms of proteins, which can be the source of novel biomarkers and are a clue to the complex enzymatic machinery located in discrete regions.
To go deeper into the precision, we performed protein imaging to link identified proteins with their precise tissue localization. This led to the localization of a C-terminal fragment of alpha-synuclein, a protein known for its involvement in Parkinson’s disease, in the hippocampal dentate gyrus.
Finally, this approach allowed the detection of novel proteins issued from non-coding regions of mature mRNA and alternative coding frames. This is called Ghost Protein issued spatial resolved ‘top-down’ proteomics associated with MALDI-MSI, which unraveled the Ghost proteome and also opened the door to the physiome in normal and pathological conditions.