Using single-cell approaches to investigate early embryonic development
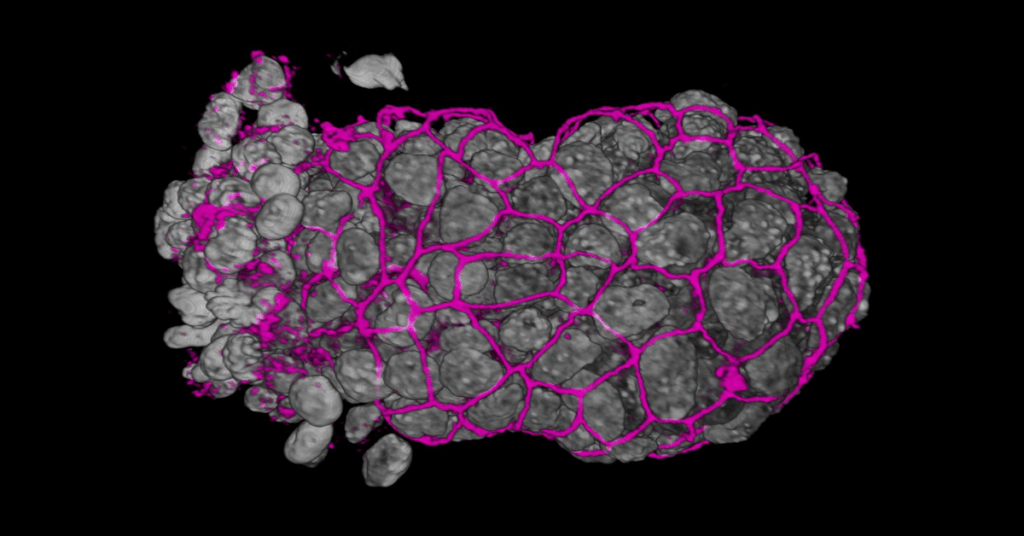
There have been rapid developments in single-cell sequencing and imaging techniques over recent years. Such approaches are particularly well suited to studying embryonic development during the earliest cell specification events. The collection of single-cell data has to go hand-in-hand with using tailored machine learning methods that drive the generation of hypotheses and the design of validation experiments.
In this webinar, we will discuss how the combination of single-cell techniques, machine learning, and developmental biology, is delivering new insights into the early events controlling embryonic development in both mice and humans.
What will you learn?
- How to analyze single-cell transcriptomic datasets taking advantage of the most recent machine learning methods
- How to generate hypotheses from sequencing data and test them with orthogonal single-cell approaches
Who may this interest?
- Those with an interest in single-cell transcriptomic data analysis
- Those with an interest in HCR data generation and analysis

Antonio Scialdone
Group Leader
Helmholtz Zentrum München (Germany)
Scialdone studied at the University of Naples “Federico II” (Italy), where he received a PhD in physics. After working as a postdoc at the John Innes Centre (Norwich, UK) in the lab of Martin Howard, and the EMBL-EBI (Cambridge, UK) in the lab of John Marioni. He established his own independent lab in 2017 at the Helmholtz Zentrum München (Germany). In his lab, he combines machine learning and physical modeling to understand cellular fate decision starting from single-cell data.
 Shankar Srinivas
Shankar Srinivas
Professor of Developmental Biology
University of Oxford (UK)
Srinivas completed a BSc in Nizam College (Hyderabad, India) before joining the group of Frank Costantini at Columbia University (NY, USA), where he received a PhD for work on kidney development. He then moved to the NIMR (London, UK), as a HFSPO fellow in the groups of Rosa Beddington and Jim Smith. Here, he pioneered the use of time-lapse microscopy of early mouse embryos to study anterior patterning. He established his own independent group at the University of Oxford in 2004, where he now uses mouse and human embryos to understand the formation of the anterior-posterior axis, gastrulation and early cardiogenesis.
This webinar was recorded on Monday 13th June 2022
Supported by
