Cancer biomarker discovery for precision medicine

Using proteomics in big data-driven precision medicine for the fight against prostate cancer.
“I got diagnosed with prostate cancer on Friday 13th. As my doctor spoke about Gleason scores, probabilities of survival, incontinence and impotence, why surgery would be good and what kind would make the most sense, his voice literally faded out like every movie or TV show about a guy being told he had cancer…,” said Mike.* “Right after I got the news, I promptly got on my computer and Googled ‘men who had prostate cancer.’ As I learned more about my disease (one of the key learnings is not to Google ‘people who died of prostate cancer’ immediately after being diagnosed with prostate cancer), I was able to wrap my head around the fact that I was incredibly fortunate. Fortunate because my cancer was detected early enough to treat and also because my internist gave me a test he didn’t have to. That test saved my life.” Three months later, after successful treatment, Mike was cancer-free [1].
Prostate cancer is the most common cancer in men and one of the most treatable cancers, if caught early enough. Statistically, one in two men aged 50 and one in three aged 30 will have prostate cancer [2]. Early detection is critical to optimize the clinical outcomes of patients. However, current screening methods result in up to two-thirds of men being over-diagnosed and thus over-treated for prostate cancer [2,3]. The current gold standard for determining the aggressiveness of prostate cancer and thus, the choice of treatment, is the Gleason Score, first developed over 70 years ago [2]. With such high over-diagnosis and over-treatment rates, it is timely to explore complementary tools to inform diagnostic and treatment decisions, to minimize the unnecessary exposure of men, with low-risk tumors that may not cause harm, to aggressive treatment with potentially harmful side effects [2,3].
Biomarker discovery for patients with prostate cancer
Therefore, the search is on for biomarkers that can better determine the level of risk with prostate cancer, as well as support precision oncology treatment decisions. While we understand the importance of a genetic component in cancer, it is a disease that can be greatly affected by lifestyle choices [4]. As such, protein – as well as genetic variants – should provide a fertile hunting ground for cancer biomarkers (see Figure 1). That is why researchers such as Tiannan Guo, Assistant Professor at Westlake University (Hangzhou, China), are investigating the proteomic profiles of samples from individuals with cancer. However, cancer samples are notoriously complex, with lots of inter-patient and inter-tissue heterogeneity, making it particularly challenging to find diagnostic biomarkers. To address these challenges, Guo and his team have developed a novel method, using liquid chromatography mass spectrometry (LC-MS) and data-independent acquisition (DIA) solutions, including SWATH Acquisition from SCIEX, for rapidly and precisely analyzing large numbers of these clinical samples to reveal their proteomic expression profiles and discover protein biomarkers to inform the diagnosis of prostate cancer [5].
This new method is a microflow, single-shot, short gradient SWATH Acquisition technique on a TripleTOF 6600 instrument. It is substantially faster than the standard proteomic strategies and thus, accelerates the discovery and verification of protein biomarkers in pre-classified clinical specimens [5]. Using a reference SWATH method with a 2-hour gradient for comparison, this new method uses only a 15-minute microflow gradient, allowing many more samples to be interrogated per day. While much faster, the new method was still able to quantify a large number of proteins (3800 across the 204 samples) with relatively high consistency (r = 0.77).
The accelerated method consumed only 17% instrument time, while quantifying 80% of proteins compared with the reference SWATH method. The 204 samples were obtained from 58 prostate cancer patients and 10 benign prostatic hyperplasia (BPH) patients. BPH is a known risk factor for developing prostate cancer. Despite missing 20% of the proteins quantified using the reference SWATH method, the 80% quantified by the new method provided similar biological information, while exhibiting 21% lower batch variation. Batch effects are difficult to avoid, resulting from technical variations in the multi-day sample preparation and LC-MS steps and are a non-trivial complication for big cohort proteomics analysis [5].

 Elaine Mardis on cancer genomics in precision medicine
Elaine Mardis on cancer genomics in precision medicine
The co-executive director of the Institute for Genomic Medicine and Professor of Pediatrics at Nationwide Children’s Hospital (both OH, USA), Elaine Mardis, delves into the techniques applied in cancer genomics and their applications in precision medicine.
Proteomic big data-driven precision medicine
Using the new accelerated method, 3800 proteins were detected, from which 75 deregulated proteins (134 peptides) were selected for further validation [5,6]. A targeted MRMHR method was then developed using the same fast gradient and instrument. To further test these 75 potential biomarkers, the ability of these proteins to distinguish benign tissues from malignant ones was tested in samples from an independent prostate cancer cohort of 154 patients. Using this second orthogonal LC-MS approach, it was shown that the proteins found were able to separate benign and malignant disease, highlighting the ability of the accelerated SWATH method to find potential markers [5]. Biomarkers discovered and verified using the methods described may one day be put into clinical use, to inform decisions regarding the determination of risk and choice of treatment for patients with prostate cancer (see Figure 2) [2,5].
Regarding the actual functions of the 75 selected proteins, they were all found to be associated with most of the pathways and metabolism networks that are significantly dysregulated in cancer. These proteins included 21 known diagnostic biomarkers, including PAP and FASN [5,7,8]. PAP, an enzyme expressed by the ACPP gene, is predominantly produced in the prostate, and is an important biomarker used to assess and monitor prostate cancer [7]. FASN, essential for tumor growth and maintenance, has also emerged as a potential oncologic target and some anti-FASN molecules have already progressed to Phase II clinical trials [7–9].
Two other proteins were also investigated in detail in the paper, PRDX3 and COPA, due to their roles in TP53 oncogene regulation, and the fact that they were both upregulated and potential drug targets [5]. The functional information not only confirms the importance of these 75 proteins identified using the new SWATH method and corroborated by the orthogonal targeted method, it highlights the applicability of the new method for the accelerated identification and quantification of proteins and peptides for biomarker discovery in larger sample cohorts. This new method may thus be an important resource for the detection of novel biomarkers for numerous diseases, beyond other types of cancer.
The discovery of biomarkers and development of new precision medicines for diseases such as prostate cancer is only possible because of the hard work of scientists such as Guo and his many colleagues in the life science research field, who are interrogating the depths of genomic, proteomic and multi-omics big data to arrive at new disease targets and treatments. Methods, such as this novel 15-minute gradient microflow SWATH LC-MS technique, facilitate the accelerated analysis of cohorts and the discovery of proteomic biomarkers and are thus helping to make precision medicine a reality.
To find out more about Guo’s research in precision medicine for prostate cancer, watch The Proteome Universe here: https://sciex.com/tpmregistration.
*Not patient’s real name
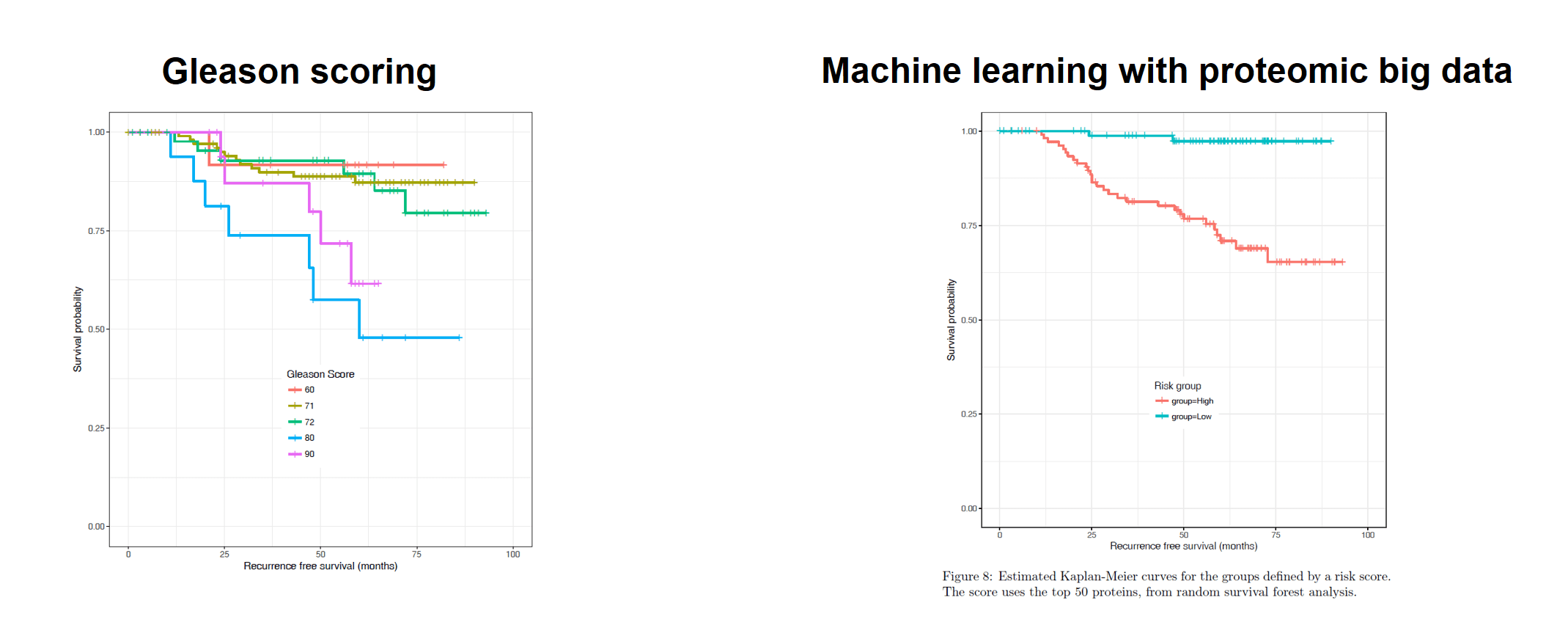
Figure 2: Potential utility of proteomic biomarkers to stratify risk for patients with prostate cancer, compared with the Gleason Score [2].
Author information
Christie Hunter is the Director of Applications at SCIEX where her and her team develop new quantitative applications using SCIEX instrumentation. They are also responsible for the technical content that demonstrates how these liquid chromatography mass spectrometry solutions can be used to solve real-world challenges across a broad range of application areas.
Disclaimer
The SCIEX clinical diagnostic portfolio is For In Vitro Diagnostic Use. Rx Only. Product(s) not available in all countries. For information on availability, please contact your local sales representative or refer to https://sciex.com/diagnostics. All other products are For Research Use Only. Not for use in Diagnostic Procedures.
Trademarks and/or registered trademarks mentioned herein are the property of AB Sciex Pte. Ltd. or their respective owners in the United States and/or certain other countries.
© 2020 DH Tech. Dev. Pte. Ltd. <RUO-MKT-19-11476-A>