Seeing SPOTS: spatial transcriptomics and protein profiling
Researchers have developed a method that can monitor cell activity throughout a tissue at high spatial resolution.
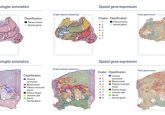
A study, co-led by researchers at Weill Cornell Medicine, NewYork-Presbyterian and the New York Genome Center (all NY, USA), was conducted to develop a method for mapping organs by combining gene activity and protein function recordings with cells’ precise locations. This method, called Spatial PrOtein and Transcriptome Sequencing (SPOTS), could be a beneficial tool for profiling cancers and identifying viable disease targets.
This study is one of many trying to increase the spatial resolution of cells in tissue, allowing for a deeper insight into organ function. Previous research has demonstrated how technologies can successfully profile gene activity; however, these methods often disregard the cells’ contextual position within a tissue. Building upon current 10x Genomics (CA, USA) technology, SPOTS provides a high-resolution spatial solution to comprehensively map tissues.
SPOTS coats glass slides used for traditional microscopy with special probe molecules, each of which has a molecular ‘barcode’ that denotes its two-dimensional location on the glass slide. When a thin tissue section is placed on the slide and permeabilized, the probe molecules can bind to the adjacent cells’ mRNAs. SPOTS can also utilize designer antibodies for targeting specific proteins of interest. Automated techniques are then used to identify the mRNAs and proteins as well as provide their spatial position in the tissue.
 A spatial approach to finding cancerous chemicals
A spatial approach to finding cancerous chemicals
Justin Colacino’s research incorporates cutting-edge single-cell and spatial techniques to explore how chemicals that we are exposed to daily change our risk of disease, such as cancer.
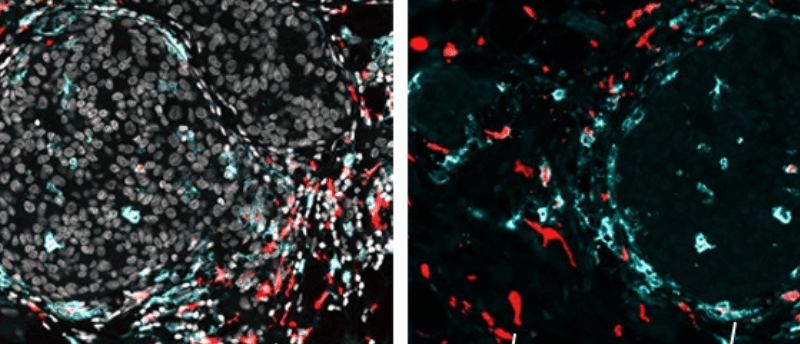
Researchers applied SPOTS to normal mouse spleen tissue to demonstrate how the method could be used to show complex tissue composition and function. They also utilized SPOTS to investigate the cellular organization of a mouse breast tumor. The resultant map indicated that macrophages, cells of the immune system, were found in two different states within the tissue: one was active and fighting against the tumor and the other had become immunosuppressive, protecting the tumor.
“We could see that these two macrophage subsets are found in different areas of the tumor and interact with different cells—and that difference in microenvironment is likely driving their distinct activity states,” reported Dan Landau (Weill Cornell Medicine and the New York Genome Project), co-senior author of the paper.
Previously too sparse to resolve, immune cells in the tumor microenvironment can now be characterized and located, an important consideration for developing cancer immunotherapies. Overall, SPOTS shows potential to increase our understanding of the tumor microenvironment and, eventually, cancer therapies.
While SPOTS’ high resolution provides extensive information about the identity of a small region of cells within a tissue, it can’t yet retrieve genetic information from a single cell. The next step, then, is to improve resolution even more to collect single-cell information.