Top tips to optimize your 3D cell culture workflows

 John Szilagyi (left) is a Principal Scientist at Bristol Myers Squibb (NJ, USA), working in nonclinical development and investigative toxicology, with a focus on the development of models to study teratology: the investigation of defects in developing fetuses and their causes. His models primarily involve the use of 3D cultures and microphysiological systems generated from multi-species induced pluripotent stem cells.
John Szilagyi (left) is a Principal Scientist at Bristol Myers Squibb (NJ, USA), working in nonclinical development and investigative toxicology, with a focus on the development of models to study teratology: the investigation of defects in developing fetuses and their causes. His models primarily involve the use of 3D cultures and microphysiological systems generated from multi-species induced pluripotent stem cells.
Here Szilagyi, reviews the utility of 3D cell models, their challenges and the solutions available to resolve them.
What is the importance of 3D cell models for the development of novel treatments and personalized medicine?
3D models allow for the efficient derivation, expansion and analysis of patient-derived tissues without the need for genetic modification to promote growth. They are superior to 2D models in that they can accurately recapitulate structural aspects of the source organ including cell–cell interactions such as those found in tight junctions.
What challenges have you encountered using 3D cell models?
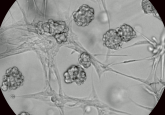
Collecting and analyzing images of 3D models is notoriously difficult to automate. Most systems are optimized to handle 2D cultures only and fail when it comes to trying to deal with the multiple and varying focal planes of a 3D culture.
What technologies and solutions did you incorporate to overcome these challenges?
The organoid module on the Incucyte® does an incredible job of capturing detailed images of entire organoid culture wells and keeping the whole image in focus. On top of that, the built-in analysis successfully separates organoids from surrounding debris and artifacts from the Matrigel to accurately determine size and darkness for a fully automated data processing workflow.
What are your top tips for obtaining the best results from 3D cell models?
Get to know your model well. Understand the growth curves, the morphological changes for death and differentiation, and the recovery from freezing and passaging. The more data you collect along the way the better, even if it doesn’t seem important. Variability is the biggest issue with organoids, and you want to make sure your assays are as consistent as possible with as many factors as you can control. Plan for extra technical replicates and seed the most organoids you can without sacrificing the quality of your assay. Even in an automated workflow or with the most trained set of hands, seeding density can be difficult to control from well to well.
What do you think the future holds for advanced cell model workflows?
I think that in the future, we should look towards analyzing the morphological changes in organoids the same way pathologists look at tissue sections. There is a lot of valuable information to be gained from observing changes to organoids under treatment beyond just size that we aren’t using yet. With advanced imaging and machine learning, it could be possible to assess organoid health in real-time without the need for sectioning and staining by simply looking at characteristics such as its shape and darkness.
This article was supported by Sartorius (Göttingen, Germany)