Molecular pencils rewrite bases

A novel base editor converts AT base pairs to GC in cells. What does this new technology offer for genetic disorders resulting from single-base mutations?

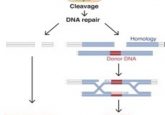
CRISPR/Cas9 technology revolutionized genome engineering by allowing researchers to edit specific DNA sequences using a simple two-component system containing a guide RNA and a Cas9 nuclease. However, the continued existence of Cas9 nuclease in cells raises the risk for cutting DNA sequences at off-target sites, thus limiting the application of CRISPR for human gene therapy .
Since a majority of human genetic disorders result from point mutations, David Liu from the Broad Institute of Harvard and MIT wondered if he could correct point mutations without creating a double-strand break in the DNA—an improvement that would reduce the possibility of introducing errors when editing genes. With this goal in mind, Liu and his team developed a base editor last year that converts GC base pairs to AT base pairs (1). Now, the team has completed their toolkit by developing base editors that convert AT to GC, which they report in Nature (2).
“CRISPR/Cas9 is like molecular scissors, while base editors are like pencils,” said Liu. “For some applications, scissors are the best tool for the job, while for other applications, such as fixing a single letter in DNA, a pencil is best.”
Enzymes that convert A to G in DNA do not exist in nature, so Liu’s team needed to evolve their own. First author Nicole Gaudelli from Harvard University created several iterations of a tRNA adenosine deaminase fused to an inactive CRISPR/Cas9 complex for targeting specific DNA sequences. The resulting enzyme converts adenine to inosine, which resembles the structure of guanine; this fools polymerases into reading it as G. In human cells, the base editor efficiently converted AT to GC with few random insertions or deletions.
Next, the team used their base editor to correct a base pair in the genome of cells derived from a patient with hereditary hemochromatosis, a disorder in which excessive iron accumulates in the body. This gene editing corrected the phenotype. They also successfully preserved a fetal gene in human cells that rescues sickle cell anemia in adults with their base editor.
“This tool could be valuable not only for in vivo therapeutic approaches but also for generating disease models,” said Kevin Holden, head of synthetic biology at Synthego, a company specializing in genome engineering solutions, who was not involved in this study.
Liu believes that these base editors can be used for correcting thousands of disease-associated point mutations by intra-purine and intra-pyrimidine transitions (A to G and T to C). In the future, Gaudelli plans to expand the toolkit further by developing an editor to correct conversions of purines and pyrimidines.