How long can bacteria tolerate antibiotics?
Measuring the minimum duration of treatment may help scientists combat antibiotic resistance by attacking pre-resistant microbes.
Our ability to treat and limit infectious disease depends on accurately characterizing pathogenic abilities. Currently, few scientists pay serious attention to signs of antibiotic tolerance, attempting instead to put out the fire of growing resistance. Biophysicist Nathalie Balaban at The Hebrew University of Jerusalem thinks it’s time for a more strategic approach.
Her team recently developed a new method to measure the time needed to kill a bacterial population completely, allowing clinicians to apprehend antibiotic-tolerant or persistent microbes before they achieve resistance (1).
“Bacteria can survive antibiotics without being resistant by arresting their growth transiently. As long as some of the population is in this state, it’s less susceptible to antibiotic action,” explained Balaban. If tolerant strains acquire even partial resistance mutations, both capabilities synergize, improving the bacteria’s survival advantage.
Although tolerance may seem like a less optimal evolutionary trait than resistance, “Tolerant bacteria can broadly survive different antibiotic treatments, but resistant bacteria may survive better under a specific antibiotic,” she said. “Once bacteria gain tolerance, they’re just a step away from resistance (2). Monitoring and detecting highly tolerant strains may help prevent antibiotic failure and relapse.”
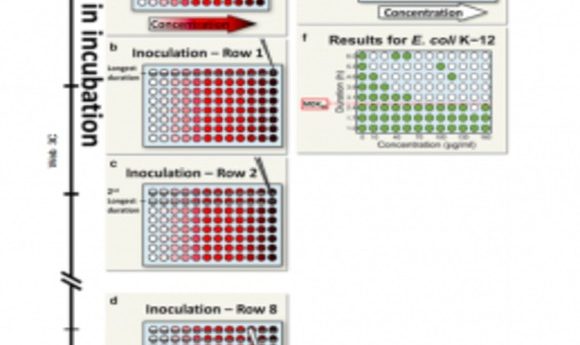
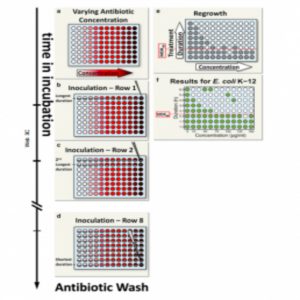
Balaban’s team proposed a simple metric for systematically assessing bacterial tolerance: the minimum duration of treatment needed to kill 99% of the population (MDK99). This protocol involves exposing populations of approximately 100 bacteria in separate microwell plates to different concentrations of antibiotics for varying periods. Survivors likely indicate tolerant strains.
Applying this technique to 6 E. coli strains revealed varying tolerance periods of 2 to 23 hours during ampicillin treatment, indicating different MDK99 values. In future studies, Balaban’s team will use this metric to study the evolution of tolerance in strains from hospital patients.
“If we wanted to follow the dynamics of tolerance for many different antibiotics and bacteria, this method would be ideal,” said population biologist Bruce Levin at Emory University, who was not involved in this study. But, “if we’re interested in following these dynamics for a few antibiotics, we’d use the conventional plating method, CFU data.”
For clinical microbiology labs, implementing MDK99 on a broader scale could provide a more efficient classification of bacterial strains and may help guide treatment decisions.