Fine-tuning synthetic protein production with CRISPR

Researchers have developed a novel method for controlling the precise amount of protein produced by mammalian cells.
A recent research effort from the Massachusetts Institute of Technology (MIT; MA, USA) has used CRISPR technology to produce a system that improves the control of protein production from cells. This system could have a dramatic impact on the production of specialized monoclonal antibodies (mAbs) for therapeutic applications and numerous other synthetic proteins.
Antibodies play a vital role in fighting disease, both in a natural immune response and as synthetically developed therapeutics, such as mAbs. In the latter application, they are produced by carefully selected and optimized cell lines. Unfortunately, there are no ‘grindset’ influencer types or self-help books for these cells to boost their productivity. Instead of playing motivational videos on a loop in front of mAb-producing bioreactors, increasing protein production efficiency requires complex cell-line development and scalability processes.
Previous efforts to promote the transcription of target genes and boost the production of the desired protein have involved the design of synthetic transcription factors, such as zinc fingers, which require a redesign for each targeted gene, making them time-consuming to develop. The MIT team aimed to address this challenge and the shortfall in transcriptional control methods by adapting the target gene and its promoter sequence.
Senior author Timothy Lu, who led this research project, had previously developed a CRISPR-based transcription factor that provided a greater deal of control over gene transcription, in 2013. The new study aimed to build on that early foundation, compiling a library of synthetic biological pieces to facilitate the delivery of transgenes alongside specifically tailored promoter regions, called “operator” sequences, that can control the level of expression from the transgene.
 AFM-IR ‘sees’ inside cells better than ever
AFM-IR ‘sees’ inside cells better than ever
A counterintuitive approach to atomic force microscope-infrared spectroscopy means researchers can study the chemical composition of human cells with unmatched precision.
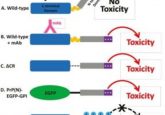
The system also includes a guide RNA that binds to the operator sequence and a transcription activation domain. The transcription activation domain is bound to a Cas9 protein that can activate gene expression upon binding to the guide RNA. The operator sequences contain between two and 16 binding sites; the more binding sites, the higher the rate of gene expression, which is how the system enables precise control of protein production. What’s more, as the binding sites in the operator sequences are distinct from naturally occurring promoters, this system is specific to the transgenes, preventing interference with native promoters and gene expression.
Finally, the team tested the system in several kinds of mammalian cells, finding positive results in Chinese hamster ovary cells, rat myoblasts, human embryonic kidney cells, and human induced pluripotent stem cells. Having demonstrated the system’s ability to produce fluorescent proteins, researchers further demonstrated its ability to produce two major sections of monoclonal antibody ‘Jug444’, but also induced Chinese hamster ovary cells to synthesize quantities of the human antibody ‘anti-PD1’. Human T cells subsequently exposed to these cells demonstrated an increased potency in tumor cell killing under conditions of increased antibody production.
While this study marks a success in terms of the antibody yield from individual cells, the process still requires significant development in order to scale it up to industrial levels. One key difference is the method of growth – while most industrially produced monoclonal antibodies are created in liquid suspension bioreactors, this process requires cells to be grown on a flat surface.
Lead author William Chen (University of South Dakota, SD, USA) explained, “This is a system that is promising to be used in industrial applications, but first we have to adapt this into suspended cells, to see if they make the proteins the same way. I suspect it should be the same, because there’s no reason that it shouldn’t, but we still need to test it.”