Flipons: the least exposed topic in molecular biology, with the most potential?

Simple changes in the DNA conformation of non-coding sequences called flipons could have a huge impact on a wide variety of diseases, including cancer, while also creating new opportunities in nanotechnology.

Alan Herbert (InsideOutBio)
When reverse transcriptase was discovered in Rouse sarcoma virus in 1970, it upended the central dogma of molecular biology that information exclusively flowed from DNA, to RNA to proteins. Its discovery vindicated the work of a series of researchers, most notably Howard Temin (University of Wisconsin-Madison, WI, USA), who had all been considered ‘oddballs,’ wasting their time on a ridiculous notion that no one else was interested in.
Years later, in 1980, building on this work and insistent on the belief that viruses containing reverse transcriptase could infect humans, Robert Gallo (University of Maryland School of Medicine, MD, USA) discovered the retrovirus HTLV-II. These two discoveries were to have a huge impact in the discovery of the virus responsible for AIDS and the subsequent strategy for its treatment.
Today a new, obscure corner of molecular biology – flipons – are under the lens of Alan Herbert, Founder of InsideOutBio (MA, USA), who is fascinated by the potential of the curious DNA structures but can’t see why others aren’t as interested in the versatile little devices. With possible applications in the treatment of cancers and a host of immune disorders, alongside various updates to nanotechnology and, could this area of study be today’s reverse transcriptase?
Please introduce us to flipons.
A flipon is a DNA sequence that can form alternate DNA conformations. Everyone knows about B-DNA, which is the traditional right-handed helix. A flipon is a completely different conformation. It can form left-handed Z-DNA, where the bases flip from right-handed to left-handed conformation. This transition is facile; you can flip from one to the other without breaking or damaging the DNA. Instead, it is a matter of energetics. If you pump enough energy into the right-handed B-DNA, you will create stress that will be relieved by flipping it from right to left-handed Z-DNA. That is called a Z-flipon.
There are two other classes of flipons. One is G-quadruplexes. A single strand of DNA that has a number of G repeats on it can be encouraged to fold into a four-sided structure that forms like a knob on the surface of DNA. The other is made from three strands, a triplex, that begins as B-DNA but then has an additional strand threaded into it to make a three-stranded structure. A lot of talented scientists have made great contributions to the knowledge base here.
The interesting thing about these flipons is that they tend to form within repeat sequences, so they contain very little information, but the sequences are ideal for flipping from B-DNA to an alternate conformation. The structures that they form are easily recognizable by cellular machinery. If a cell has an element of its three billion base-pair genomes that it wants to highlight for an action, a flipon is ideal because it stands out against all the B-DNA.
The other interesting thing about flipons is that you can change how easily they flip from one conformation to another either by DNA modification, with small molecules or even by applying physical force to alter the DNA twist or even its length.
What is the natural function of flipons?
Z-flipons are involved in regulating the immune response – there is genetic evidence for this – by associating with the RNA-specific enzyme ADAR (adenosine deaminase). RNA transcribed from repeat sequences can fold onto themselves, forming double-stranded RNA (dsRNA). ADAR edits this RNA as it is transcribed, changing the information content that’s read out from DNA, destabilizing the dsRNA. This is important as dsRNA has several negative implications for a cell. For example, when dsRNA is formed in a transcript, it can hide the signals needed for correct splicing of a mRNA, preventing incorporation of an exon into the final message. By destabilizing the dsRNA, editing can make these sites available for use. The formation of Z-DNA in the repeat sequences is driven by their transcription and can vary with DNA modifications and context. It localizes ADAR to these sequences.
In addition to effects on splicing and recoding of mRNA, editing prevents unnecessary cell death due to abnormally high levels of dsRNA. Cell sensors exist to detect this situation. Usually they protect against virus infections where dsRNA is produced in excess by initiating cell suicide or elimination of the cell by the immune system. . The ADAR system is used by cancer cells to protect themselves as they lose control of transcription and begin to produce a lot of dsRNA.
The G-quadruplexes are highly enriched in areas that control transcription of genes and their formation is associated, in many cases, with increased readout of DNA repair enzymes. G-quadruplexes are easily induced or modified by chemicals and mutagens, so DNA damage can favor their formation. The persistent formation of the quadruplex indicates that the cell is under stress and needs to amp up its production of DNA repair enzymes, so they provide another example of a flipon protective mechanism.
Why did we evolve to contain these sequences?
Essentially flipons arose because of the threat of junk DNA, which would just copy and paste itself throughout the genome. Junk DNA does not encode any protein, it simply replicates itself. In humans there is a series of sequences called ALU, that were rampant in the genome early in human origins. These sequences were problematic because they tend to target very active genes, causing mutations and dangerous random effects. They were an existential threat to humans in the early stages of our development and proteins like ADAR stopped their spread.
Initially, these repeats were recognized due to the fact that they formed these alternate DNA structures. By latching onto the Z-flipons and modifying the RNA, it prevented the sequences being copied back into the genome.
Flipons and their interactions developed to manage the threat of these invasive sequences but, as often happens with invasive components, over time the invader became assimilated and got put to work. Flipons became a way of highlighting genes to have their RNA edited so that from one gene you could produce multiple different transcripts. It became a mechanism for increasing phenotypic diversity and targeting cellular machinery to particular sites in the genome. Some of these machines cause histone epigenetic modifications providing a more nuanced way to regulate gene expression.
You have mentioned that flipons can impact cancer biology, what is the nature of this interaction?
As mentioned, the repeat sequences, especially ALU, can form an opposite orientation that, when read out into RNA, can form dsRNA. Once dsRNA in the cells goes above a certain threshold it triggers apoptosis. If the cancer cell mutates to avoid apoptosis then the dsRNA acts as a signal to the immune system that the cell is dangerous and needs to be eliminated.
Cancer cells, due to the characteristic excessive transcription, end up transcribing so much dsRNA that they exceed the safe threshold and are in danger of either apoptosis or immune attack. To combat this, they over-express the enzyme ADAR that recognizes the Z-flipon to try and get rid of the dsRNAs.
What could this mean for cancer therapies?
The traditional thinking of cancer drug treatment is based on codons: DNA mutation leads to cancer, find the DNA mutation, find the product of that gene and inhibit that mutated protein. That is the way people thought for 50 or 60 years. In many cases it has led to life-saving treatments. With flipons in cancer, the focus is on non-coding RNA, not DNA. By inhibiting ADAR, immunotherapy works better as the alarm signals due to the abnormal formation of dsRNA now function. It is a completely different axis and it is something that no one had imagined we could work with, even five years ago.
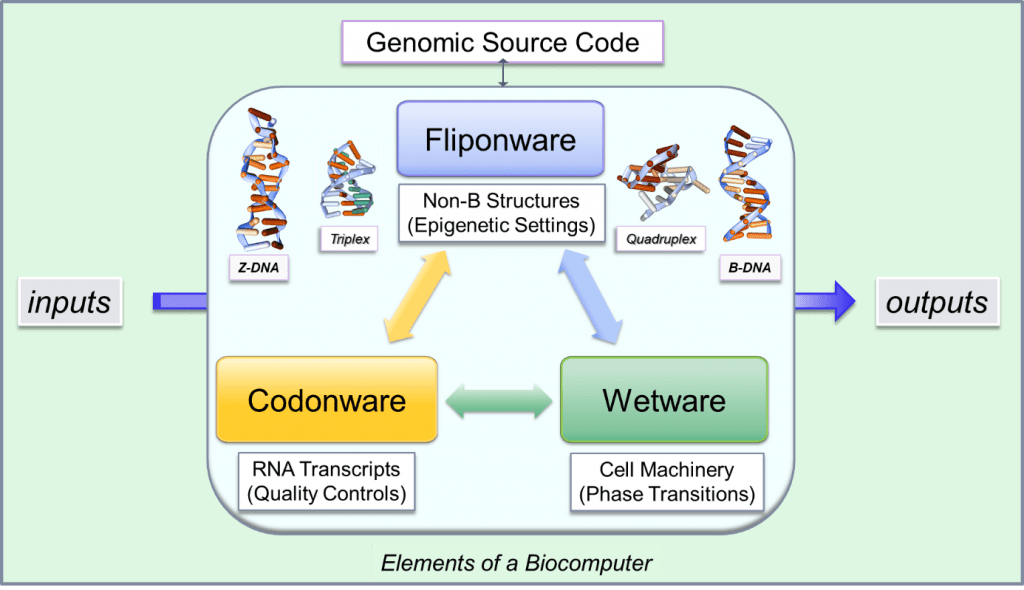
A schematic of fliponware and its interaction in DNA computers.
Your recent papers look at the use of flipons in DNA computers. Can you introduce the concept of DNA computers?
In simple bacterial genetics, you have a very small genome and you can identify clearly which protein will be expressed from a specific DNA sequence. This is a simple DNA computer; the software is based on codons – or codonware – and the protein is the wetware. In larger human genomes, flipons allow a higher degree of complexity. They enable the production of multiple proteins from a single gene. I refer to this system as fliponware.
With multi-cellular organisms, looking at the DNA sequence in most cases only allows you to map possible protein products, but not necessarily those that are important to a particular outcome. Nor does sequence gazing allow you to know how flipons affect this process. Nature has used the variability provided by flipons to evolve complex genetic programs that can switch quickly from one output to another.
With fliponware you can get many different transcripts from a single gene. The flipon is analogous to a software compiler, allowing you to assemble the code blocks in different ways to produce a more intricate machine code. Essentially, codonware represents the traditional genetic code Fliponware increase the number of ways these codon blocks can be combined. It allows you to run different genetic programs with the same set of genes, all without changing the DNA sequence.
Fliponware can be controlled and influenced by proteins. For instance, if you methylate Z-flipons to form sequences in a certain way, you can lower the threshold of forming a Z-flipon, changing the ease with which they form. It’s a very dynamic code. It gives you a huge dynamic range in terms of which RNAs you make and it allows the body to select out cells that survive best given the current circumstances.
What are some of the applications of this fliponware in the life sciences? What do you think that it can begin to achieve?
Flipons are already in use in lots of laboratories around the world. They have been used to make biosensors to try and understand the conditions that cause B-DNA to swap to an alternate conformation. The exciting thing is that these are incredibly small biosensors. If you look at the limits of silicon technology, it’s around 7 nm now. But if you look at the diameter of a G-quadruplex, it’s 0.025nm. These elements flip from one state to another in different conditions. This allows you to make reporters out of them easily and in different ways. You can use fluorescence resonance energy transfer (FRET) to get a signal out of them. You can also use the flipon structure to stabilize DNA enzymes or RNA enzymes in an active conformation. Once the flipon forms, the DNA enzyme is active. It will then catalyze a reaction and produce a product that you can sense as the readout. For example, hemin has already been used with G-quadruplexes to form an artificial DNA catalase.
Flipons can be manipulated to influence RNA editing in certain situations to create start or stop codons in genes. This allows you to create cells that detect when a specific event or environmental change has occurred. For instance, if a mutagen is present that causes the DNA to flip conformation, you could then report its presence via the flipon dependent expression of a protein that leads to production of a reporter gene. The signal then could be further amplified by a flipon that promotes cell growth, making exponentially more copies of the reporter.
Another key feature of flipons is that they are sensitive to DNA tension. If you use optical tweezers and pull on DNA, you can cause enough stress in the DNA to flip it from B to an alternate conformation. This means that you could put the flipon system onto a flexible membrane and use it to report very accurate distance measurements, using an optical sensor to detect when the flip occurs.
DNA helices can conduct charge and flipons normally introduce a kink into the DNA. You can use that kink to make a contact across a junction, producing a gate. Eventually it may be possible to make a very small micro-switch to bridge a junction, mimicking the way semiconductors are used in transistors but on a much smaller scale.
G-quadruplexes offer the potential for digital coding. When they fold up into their quadruplex, they are very stable. This enables them to be assigned a 0 when they are B-DNA and a 1 when they are in a G-quadruplex. Their formation is completely reversible, you could erase the G-quadruplex by changing the ionic conditions or by using heat or by light induced isomerization. You can go from 0 to 1 and back to 0 in a controlled fashion without changing the DNA sequence.
There is a range of nanopore technologies in which you could use flipons. For example, with a single-stranded nanopore, you could regulate whether or not the DNA goes through that pore by forming a G-quadruplex which is too big a structure to fit through the pore, blocking it. There are ways of sensing whether that pore is open or not. So G-quadruplex formation is a very compact way of actually having a 1 or a 0 state that you could develop into a useful device – memory storage, for example, or to act like the tape used in a Turing machine.
What got you into working on fliponware? And what do you love about this area of science?
Science is fun because there are lots of problems that nature has solved that we really do not understand. It is always exciting to find a solution that nature has come up with which, in retrospect, is both brilliant and remarkably simple. I think everyone looks at nature as being a little bit sloppy, a little bit opportunistic, but it’s actually a great system for finding simple solutions that are later elaborated into complex outcomes. That excitement of science is always something that’s been with me and life is too short to work on easy problems. I know working on difficult problems is risky and stressful, it can be tricky to get funding and people do not talk to you because you are bit of an oddball, but then there is the thrill of discovery.
So that desire to understand the ways nature has solved certain issues led me to study Z-DNA and flipons. While it didn’t have an immediate application, the Z-DNA work ended up having great relevance to cancer and to a subset of immune diseases, so it’s really exciting when something that’s driven basically by curiosity ends up having highly impactful outcomes in medicine. I am sure once the ADAR inhibitors get into clinic, paired with their checkpoint inhibitors, then we will see another huge step forward in cancer treatment because this represents a different axis of treatment.