Hidden microbial treasures
A new study shows that most of the human microbiome consists of unidentified microbes.

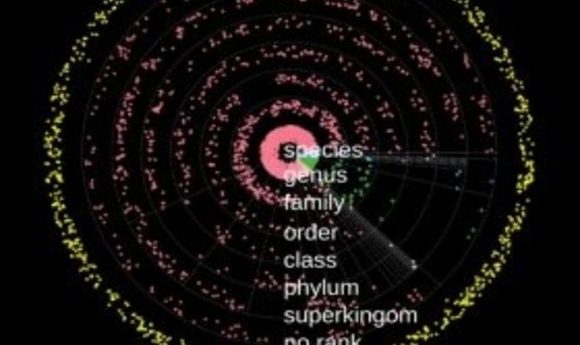
Solar system plot showing new overlapping sequences greater than 1 kbp in size. Rings represent different taxonomic levels, while the radius of each point in the ring indicates the average gene homology. Gray dotted spokes separate the superkingdoms or phyla. The yellow dots represent sequences unassigned to any superkingdom. Different colors indicate different superkingdoms; counterclockwise from 3 o’clock: bacteria, viruses, eukaryota, and archaea.”
Credit: Mark Kowarsky, Stanford University.
Over the last decade, the number of publications linking an individual’s microbiome to health problems ranging from migraines, to diabetes, to Parkinson’s disease has increased 10-fold. Given this progress, it was easy to believe that the majority of microbes inside the human body had been identified. Surprisingly, a recent study published in Proceedings of the National Academy of Sciences reports that a whopping majority of the microbes comprising our microbiomes remain unknown (1).
“It was kind of just a lucky discovery,” said Mark Kowarsky from Stanford University, lead author of the paper. Previously, Kowarsky’s colleagues developed a non-invasive method for detecting organ transplant rejection by patient immune systems by characterizing circulating cell-free (cf) DNA from blood samples (2). This cfDNA includes the patient’s own DNA, DNA from the organ donor, and non-human DNA from microbes and viruses.
While analyzing the cfDNA, the team noticed something unusual — 99% of non-human DNA fragments circulating in our blood stream did not match any of those in the existing databases of known microbes and viruses. Intrigued, Kowarsky set out to characterize these unidentified microbes.
In their new study, Kowarsky’s team obtained more than 1000 blood samples from immuno-compromised humans, including organ-transplant patients and pregnant women, since pregnancy also alters immune function. Then, they assembled the new sequence data without a reference genome, to classify the microbes. They found hundreds of new bacteria and viruses that were previously not known to be a part of the human microbiome. Some of these microbes belonged to completely new taxa, implying that researchers previously underestimated the diversity of the human microbiome.
“It’s very exciting that they have seen so many things we have never seen before in other studies,” said Elisabeth Bik from uBiome, a company specializing in microbial genomics.
These microbes may have eluded detection up until now because conventional microbiome studies assessed microbes from select body parts such as the gut or skin. Blood samples represent the entire body. Likewise, researchers have studied cfDNA for years, but most of these studies focused on human cfDNA, neglecting the circulating non-human sequences.
This study relied on DNA from immuno-compromised patient blood samples, but in the future, Bik is curious about what researchers could find in the blood of healthy people. “This study shows there’s a lot more to be found,” she said. “We are not there yet.”