DNA nanostructures manipulate cell membranes for optimized drug delivery
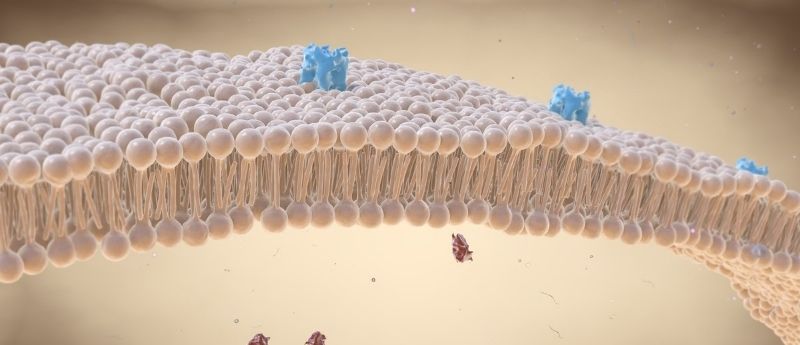
By creating DNA origami nanostructures to manipulate lysosome cell membranes and optimizing the conditions for their binding, a team of researchers aims to improve their drug delivery and biosensing capabilities.
A recent research effort led by Matthew Baker (University of New South Wales, Sydney, Australia) and Shelley Wickham (University of Sydney, Australia) has identified the optimal process to manipulate DNA ‘nanostructures’ to interact with cellular membranes, such as lysosomes. The study could lead to the development of small molecular systems that could dramatically improve the fields of biosensing and drug delivery.
The use of liposomes, nanosized ‘bubbles’ of cellular membranes, has been well established in drug delivery for cancer and other diseases. However, there is still much room for improvement in this delivery of therapeutics and recently researchers have been using DNA nanostructures to modify the porosity, shape and reactivity of a liposome to deliver this improvement. Getting the DNA nanostructure to effectively interact with the lipid membranes has been a challenge in this endeavor; a challenge that Barker and Wickham set out to resolve.
The team identified that two key issues were obtaining the best buffer conditions to facilitate binding of the DNA to the liposomes and establishing the optimum amount and position of cholesterol additions to the DNA nanostructures that enable them to bind to liposomes.
To address this, the team characterized the effects of each of these conditions including DNA structure, cholesterol conformation and buffer and lipid composition on DNA-liposome binding. The optimal conditions for each factor are documented in the study; For instance, the ideal number of cholesterol modifications to bind a 2D DNA origami tile is between 4 and 8, while placing the modifications on the edge of the tile and using a greater linker length between cholesterol and tile both improved binding.
With the best conditions for binding established, the team demonstrated the increased capability of the optimized liposomes. Describing their achievement, Marker revealed that the team has built a, “…totally new DNA nanotechnology where we can punch holes in membranes, on demand, to be able to pass important signals across a membrane.” This clearly demonstrates the advantage that this research could provide for therapeutics that need to deliver their active molecule to a very precise location before releasing them.
However, the impact does not stop with drug delivery. The authors also note that by imbuing lysosomes with the ability to detect their environment new potential applications present themselves.
“One major application of our study is biosensing: you could stick some droplets in a person or patient, as it moves through the body it records local environment, processes this and delivers a result so you can ‘read out’, the local environment,” notes Barker
The authors hope that the research can be used to design more complex lipid-binding nanostructures that can be used for a broad range of applications within nanomedicine, nanotechnology and nucleic acid research. They also intend to develop methods to control DNA pores with light, essentially developing synthetic retinas.