Building the human brain cell atlas

In an international collaboration, researchers hope to integrate single-cell and spatial techniques to create a cell atlas of human and primate brains.
A cell atlas refers to a comprehensive map and profile of each cell present in a tissue. Now, biotech companies and researchers are interested in developing technologies that can determine not only cell type and location, but also shape and distribution of human and primate brain cells. Presented at the 2022 Society for Neuroscience Annual Meeting (San Diego (CA, USA); 12–16 November 2022), these methods for developing a brain atlas represent the advancement of spatial visualization techniques for the monitoring of brain changes and disease.
Previous work has developed a cell atlas for the mouse brain, which contains approximately 100 million cells. However, human and primate brains have billions of cells, making the creation of a cell atlas much more complex. Five research groups around the world are developing and utilizing different spatial and single-cell technologies to visualize brain cells at high resolution in context, often applying them first to mouse models before hopefully transitioning to humans.
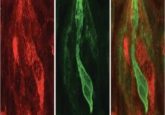
Researchers from the French National Center for Scientific Research at the University of Bordeaux (France) are augmenting super-resolution shadow imaging (SUSHI) with 2-photon shadow imaging (TUSHI) to investigate the mouse brain in vivo. SUSHI captures the extracellular space around brain cells, allowing for the visualization of brain tissue at nanoscale spatial resolution. However, combining SUSHI and TUSHI provides more information about mouse brain anatomy in vivo [1].
At the biotechnology company Vizgen (MA, USA), scientists are using multiplexed error-robust fluorescence in situ hybridization (MERFISHTM) to create a mouse brain atlas that is both spatially resolved and molecularly defined. This technology demonstrates how spatial genomic assays allow for molecular-level tissue analysis [2].
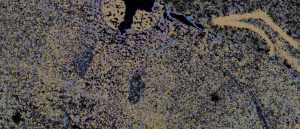 Navigating gene expression with MERFISH – the Google Maps of biology
Navigating gene expression with MERFISH – the Google Maps of biology
We spoke with George Emanuel from Vizgen about advancements in fluorescence in situ hybridization and the development of MERFISH.
A transcriptomic mouse brain cell atlas, built by scientists at the Allen Institute for Brain Science (WA, USA), integrated a number of whole-brain single-cell RNA-sequencing (scRNA-seq) datasets. They then annotated these datasets using MERFISH to mark the location of each cell type. From here, the researchers created a comprehensive distribution map of neurotransmitters and neuropeptides to enhance the characterization of cell types throughout the mouse brain [3].
Akoya Biosciences (MA, USA) have developed the PhenoCycler-Fusion platform that uses an advanced imaging technique, multiplexed immunofluorescence imaging, to clearly and precisely visualize hundreds of biomarkers in a tissue. This single-cell, subcellular resolution imaging technique is currently being used to compare the neuronal populations of healthy and diseased brains [4].
Finally, clear, unobstructed brain/body imaging combination and computational analysis (CUBIC) methodology has been developed by the RIKEN Center for Biosystems Dynamics Research (Kobe, Japan), which has been used to locate 400 million cells in a newborn mouse’s whole-body atlas. Additionally, CUBIC’s tissue clearing abilities have allowed researchers to view the entire adult mouse brain with single-cell resolution [5].
“Ultimately the different data sets generated by different methods can be integrated together to provide a comprehensive description of the different properties of the cells that can go into the same cell atlas,” commented Hongkui Zeng, director of the Allen Institute for Brain Science. “Ideally, in the end, we’re all converging into the same system to make a holistic, complete cell atlas.”