A better way to profile breast cancer
A new single-cell RNA-seq transcriptome profiling technique allows researchers to quickly characterize the wide variety of cells comprising tumors, along with neighboring stromal and immune cells in the tumor’s microenvironment.
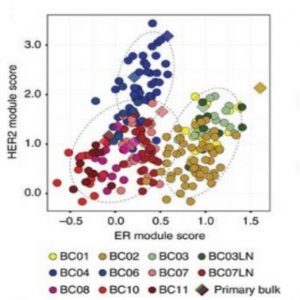
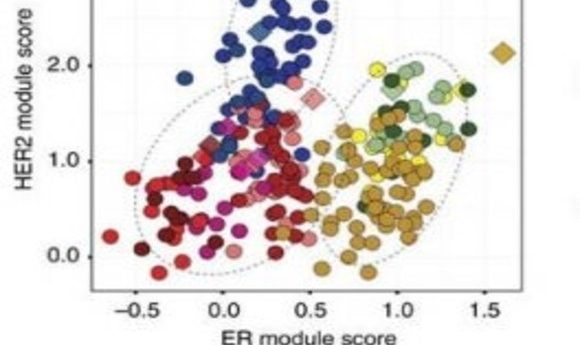
Subtype prediction of tumour cells (1).
In the past several years, molecular targeted therapies have significantly improved outcomes for breast cancer patients. Clinical laboratory personnel normally use genomic and gene expression profiling to characterize bulk tumors in individual patients, but cancers typically display significant heterogeneity within the tumor that might affect treatment efficacy. Single-cell transcriptome analysis of circulating tumor cells can provide a more precise estimation of tumor heterogeneity, possibly leading to more effective targeted therapies.
As recently reported in Nature Communications, Woong-Yang Park from the Samsung Genome Institute in South Korea and his research team analyzed the transcriptomes of 515 single cells from 11 patients with four different breast cancer subtypes: luminal A; luminal B; HER2; and triple negative breast cancer (TNBC). Inferred copy number variations (CNV) from the single-cell RNA-seq data helped the researchers identify distinct cancer populations, each characterized by a unique pattern of gene expression. For example, the team discovered that TNBC cells expressed more aggressive cancer signatures such as genes involved with epithelial–mesenchymal transition and angiogenesis.
“We also wanted to know the complexity of immune cells infiltrating tumor tissue,” Park said. Each tumor manifested a differential level of immune cell infiltration; for example, luminal A type tumors were highly enriched with cancer cells, whereas most TNBC type tumors showed extensive immune cell infiltration.
Occasionally, tumor-infiltrating T cells get naturally turned off as a way to prevent overly aggressive immune responses. These T cells display a so-called “exhaustion profile.” Drugs called checkpoint inhibitors have the potential to turn these T cells back on in order to better fight cancer. Park and colleagues’ single-cell technique revealed TNBC tumors that carried “exhausted” T cells.
“Finding these T cells in there is pretty exciting because they give a clue as to why the cancer got out of control and also present a way to target tumors therapeutically,” said John Beausang, a genomicist at Stanford University who was not involved with the study.
Researchers traditionally have used immunostaining to assess the population of immune cells. However, assessing CNV is less time consuming, giving the new technique a quicker turn-around time for potential clinical applications.
Park and his team plan to focus next on using their single-cell technique to profile other cancer types such as lung cancer.