Ribosomal DNA and cancer
What can changes in ribosomal DNA, an often-overlooked part of the genome, tell us about cancer?
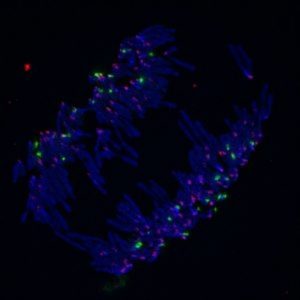
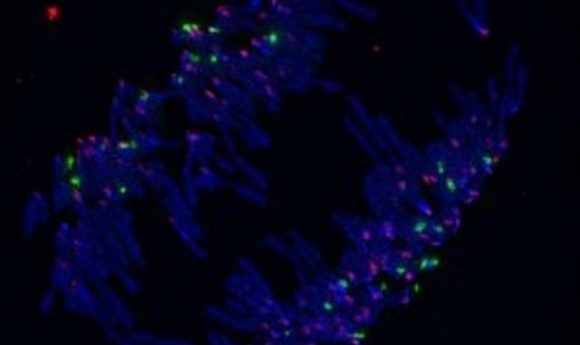
DNA (blue), rDNA (green), and centromeres (pink) labeled in anaphase.
Credit: Tamara Potapova from the Gerton lab
Ribosomal RNAs form core structural and functional components of the ribosome, but despite their central role in protein synthesis and their abundance in cells, little is known about the ribosomal DNA (rDNA) that encodes them. In humans, there are two rDNA genes: 5S and 45S. The 45S rDNA encodes a single transcript that is processed into separate 28S, 18S, and 5.8S rRNAs; it exists as several copies, or repeats on the five acrocentric chromosomes. In contrast, 5S rDNA resides on chromosome 1.
While both rDNAs are transcribed rapidly to meet the protein demands of proliferating cells, only about half of the 45S repeats are transcribed, and the repeats exhibit considerable copy number variability (CNV) among individuals. Yet, genome sequencing studies have largely overlooked these variations and their potential significance in human disease. Now, Jennifer Gerton at the Stowers Institute for Medical Research in Missouri and Bernardo Lemos at Harvard University have identified significant changes in rDNA that occur in tumor cells.
From Model Organisms to Humans
A few years ago, Lemos teamed up with Keith Maggert at Texas A&M University to investigate rDNA CNV in Drosophila (1). “In fruit flies, we saw gene expression changes associated with deletions in rDNA copy number, and there were other phenotypic consequences that Keith observed as well,” Lemos said. “We and others had documented extensive natural variation in rDNA (2) and also showed that this was associated with gene expression changes in hundreds of genes across the human genome.”
This finding sparked Gerton’s interest in exploring the relationship between rDNA CNV and cancer. Her team first confirmed that they could precisely measure rDNA copy numbers in different mouse tissues using droplet digital PCR(ddPCR) and compared their results with the changes observed by Lemos (2) to confirm that their approach was sufficiently sensitive to detect copy number changes in different tissues.
Next, they analyzed whole-genome sequence (WGS) data for tumor–normal pairs from 162 individuals representing 8 different cancer types along with WGS data from 143 healthy individuals (3). While the numbers of 45S rDNA repeats from normal tissue varied, the group noted a significant decrease in 45S rDNA repeats for three of the eight cancer genomes (osteosarcoma, AIDS-related lymphoma, and esophageal adenocarcinoma) (3).
“We know that in yeast, rDNA copy numbers can expand or contract. In cancer, we thought they would expand, so we were surprised to see that they contracted,” Gerton said. However, the team also found that copy numbers of other genes, especially those coding for metabolic and DNA repair proteins, increased in tumor cells, suggesting that a loss of rDNA copies may accompany global effects on the genome.
In a similar study, Lemos’ group examined WGS data from 721 tumors and 225 adjacent normal tissues representing 6 cancer types. While they observed slight but significant decreases in copy numbers for 45S rDNA in five of the cancers, they also noted a significant and striking increase for 5S rDNA in four of the cancers, with lung adenocarcinoma showing the greatest decrease in 45S and increase in 5S. They attribute the gain of 5S rDNA in these cancers to both internal chromosome duplication in the region that contains the 5S gene and local expansion of the gene itself (4).
An Open Question
To investigate how reductions in rDNA copy number might contribute to oncogenesis, Gerton’s group examined the relationship between rDNA loss and mTOR, a master regulatory kinase that couples cell proliferation, protein synthesis, and nutrient availability and is activated in many cancer types, including those they examined that showed a loss of rDNA (3). They observed a reduction in 45S rDNA copy number in hematopoietic stem cells (HSCs) from mice mutant for Pten, a negative regulator of mTOR, whose loss of function occurs in many cancers. When compared with tissues from the same mice that had normal Pten, Gerton and her colleagues noted an increased sensitivity of the Pten mutant cells to DNA damaging agents (3).
Meanwhile, Lemos and his colleagues surveyed their data for mutations in other genes besides those for rDNA and found that mutations in the gene coding for TP53, another protein with well-known roles in cell proliferation and DNA repair, correlated with a reduction in 45S copy number for half of the cancer types.
How rDNA might interact with PTEN, TP53, and other proteins to influence cancer remains unknown. “My conservative hypothesis is that [rDNA changes] are not a cause of oncogenesis,” Gerton said. There could be other sequences that contract, and the cells that have these contractions could have a competitive growth advantage.”
Gerton’s group went on to show that DNA replication stress in cancer cells selects for a lower copy number, making the genome easier to replicate (5). Based on these results, Gerton thinks that rDNA may one day serve as a diagnostic marker for replicative stress, allowing physicians to target tumor cells with DNA-damaging agents.
Much remains to be learned about the relationship between rDNA and cancer. “[This research] is only a small step really.” Lemos said. “Cancer is a complex and tricky disease, and the rDNA repeats have been a black box for far too long. These findings open novel possibilities to target cancers.”