The uncertain future of CRISPR Cas9 technology
CRISPR-Cas9 technology breathed new life into gene therapy as basic science and pre-clinical studies indicated early success, but researchers now doubt whether it will translate into the clinic.
Gene editing with CRISPR-Cas9: Cas9 (peach) cleaves DNA at specific sites with the help of guide RNA (grey).
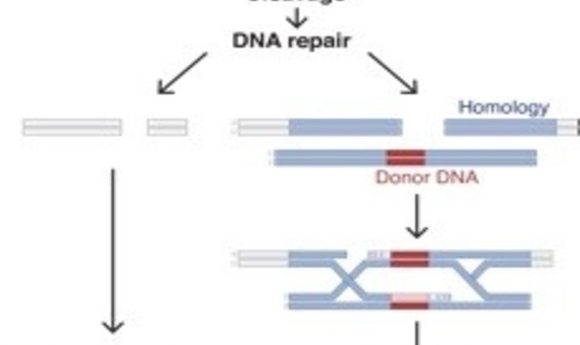
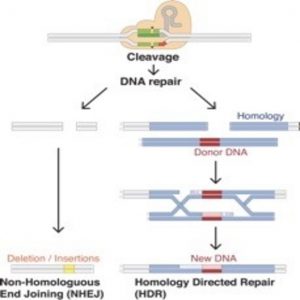
In theory, the concept of editing the genome to correct for abnormalities seems straightforward, if ambitious. Scientists have tried for decades–going back to the bubble boy syndromein the 90s–to apply gene editing for therapy with varying degrees of success. The biggest hurdle lies in the delivery system, often a viral vector, which poses a risk for host gene integration or mutagenesis.
In 2013, researchers showed for the first time that CRISPR-Cas9 could be used to edit the genome in eukaryotic cells (1), offering a great deal of control over where the editing occurred. Since then, scientists have improved the efficiency of this system, created highly specialized transgenic mouse models, and corrected many genetic disorders. In vitro and in vivo results all pointed toward clinical success. But now a new study by Matthew Porteus at Stanford University suggests that the success of this system in basic science may not translate into clinical gene therapy because humans possess pre-existing immunity to bacterial Cas9 proteins (2).
The Ghost of CRISPR Past
Inventions often arise from unrelated endeavors. For example, while working with a microwave-emitting magnetron, engineer Percy Spencer realized that the radiation was causing a nearby chocolate bar to melt. He introduced the world to the microwave oven not long after that.
CRISPR-Cas9 also resulted from an accidental finding in 1989, born from curiosity when then graduate student Francisco Mojica came across copies of palindromic repeat sequences regularly separated by spacers in the genome of a salt-tolerant microbe, Haloferax mediterranei (3). The regularly interspaced repeat sequences were unlike any family of repeats known in microbes.
These structures were soon identified in many more microbes, and pioneers in the field coined the term CRISPR to represent these Clustered Regularly Interspaced Palindromic Repeats. After observing the repeat sequences in bacteriophage genomes, researchers proposed that these sequences helped provide immunity in microbes against bacteriophages, and in 2007, scientists Rodolphe Barrangou and Philippe Horvath from Danisco A/S showed this to be true.
Barrangou and Horvath showed that the bacterial genome accommodated parts of the bacteriophage genome as a defense mechanism, preventing further attack by bacteriophages in Streptococcus thermophilus (4). Piece by piece, researchers elucidated how CRISPR-Cas9 played a role in microbial immunity. CRISPR sequences bookend short stretches of DNA that bacteria have essentially copied from invading phages, preserving a memory of the attackers. The CRISPR sequences are then transcribed into short guide RNAs (gRNA), and the gRNA then guides the Cas9 nuclease to any matching phage DNA. Cas9 cleaves the foreign DNA, inactivating it.
The 2013 demonstration of CRISPR-Cas9 editing genes in eukaryotic cells introduced a superior way to manipulate the genome in vitro and in vivo, reviving enthusiasm for the possibility of using gene therapy to cure diseases. Gene therapy has generated interest for decades, but has had difficulty delivering on its promises because of many hurdles involving targeted gene delivery and activation and the host immune response. CRISPR-Cas9 addresses most of these problems, but it is not yet clear whether it will be clinically effective.
The Ghost Of CRISPR Present
The human immune system is complex, and despite extensive study, researchers still cannot precisely predict the immune response to any foreign or self-antigen, cell, or microbe. Time and time again, the immune system proves responsible for the failure of treatments introduced using DNA, microbes, or cells: graft-verus-host disease, immune rejection of cells, severe immune response to viral vector proteins. As a result, patients with pre-existing antibodies against either the vector or the transgene must be excluded from clinical gene therapy trials to prevent an adverse immune response and in severe cases, death.
Porteus was intrigued by the translatability of the CRISPR-Cas9 system in humans, as researchers most commonly used Cas9 proteins isolated from bacteria that are human pathogens, Staphylococcus aureus (S. aureus) and Streptococcus pyogenes (S. pyogenes), for in vivo gene editing. It seemed likely that humans possessed immunity against these pathogens. “We didn’t see anyone following up on that [immunity] and we decided to look ourselves,” Porteus said. “Because we had access to serum from adults and we had Cas9 proteins, we wanted to see if healthy adults have antibodies that recognize these Cas9.”
When they began, Porteus’ team didn’t expect to see a detectable immune response since Staphylococcus and Streptococcus species often exhibit multiple antigens on their surface and Cas9 is an intracellular protein. The team collected healthy adult and cord blood, mixed it with the 2 different Cas9 proteins, and assessed the presence of antibodies. (Although derived from newborns, cord blood carries antibodies passed through the placenta from the mother, providing passive immunity to the newborn.)
Porteus’s team was very surprised by the results. “We saw that in fact, 70% of healthy adults actually have antibodies to both types of Cas9. That sort of opened our eyes, like ‘Oh it’s real!’ So then we thought, antibodies are one arm of the adaptive immune response, what about T-cells, which might be more problematic?” recalled Porteus.
Adaptive immunity consists of B-cells and T-cells, and this arm of immunity is often most effective in fighting off infections. When stimulated with an antigen, B-cells produce antibodies specific to the antigen, while T-cells can either complement B-cell production of specific antibodies (helper T-cells) or develop into killer T-cells that recognize any cell that expresses this specific antigen.
To find out if immature T-cells in adult blood and cord blood could be stimulated with Cas9 proteins to become helper or killer T-cells, the researchers mixed the appropriate fraction of blood containing immature T-cells with Cas9 proteins. The immune system of the newborn turned out to be incapable of mounting an immune response, but the team was fascinated to learn that Cas9 from S. aureus induced a T-cell response in adult blood. The stimulated helper and killer T-cells were antigen-specific. This finding raises a red flag for gene therapy trials since administering Cas9 from S. aureus could cause a serious inflammatory response and tissue death in a clinical setting, said Porteus.
The Ghost Of CRISPR Future
“It’s still very early days,” Porteus said. “We don’t think that this means the excitement of the technology is decreased in any way, but we do think that it raises an issue that needs to be considered.”
In this case, excluding patients with pre-existing immunity to the vectors or transgene as done in previous clinical gene therapy trials would not be helpful; Porteus expects that the majority of the population has pre-exisiting immunity to Cas9. But researchers certainly aren’t going to give up on their efforts to develop effective gene therapies for clinical use.
“I think there are a number of things that people will do: 1) Derive Cas9 systems from other bacteria that do not infect humans, 2) look at when in life immune responses develop to S. aureus, as this system could work in children, and 3) identify which regions of the protein are more immunogenic,” said Porteus. “I think it is possible, but I think it is challenging. The human immune system is very good.”
Porteus hopes his work will encourage researchers to factor in the immune response while considering the effectiveness of a technology. “The human immune response has consistently caused problems for gene therapy and I think it’s good that the genome editing field learns about this. Understanding it more will help figure out ways to get around it,” remarked Porteus.