Giant viruses among us: enlarging the domain of viruses
Viruses are considered the tiniest pathogens, rarely exceeding 150nm in diameter. However, the discovery of giant viruses cast the origins and nature of these infectious agents into question.
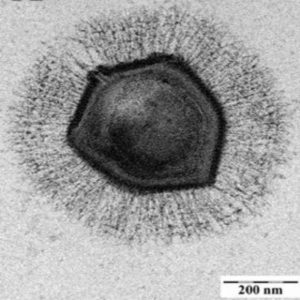
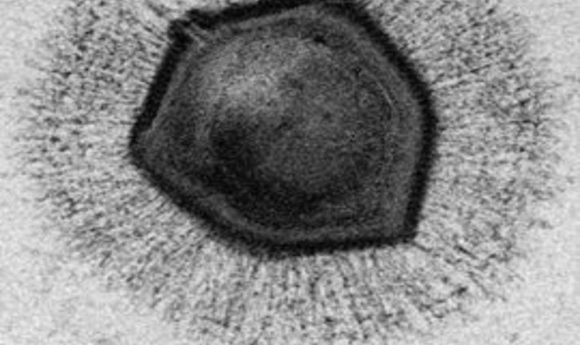
Electron microscope image of a mimivirus.
Credit: Wikimedia
In 1892, Russian botanist Dmitry Ivanovsky made the surprising discovery that extracts from tobacco leaves remained infectious even after passing through a Chamberland filter—a porcelain filter believed at the time to exclude all living microorganisms. Six years later, Dutch microbiologist Martinus Beijerinck first coined the term “virus” when he described these filterable infectious particles, calling them “contagium vivum fluidum”—contagious living fluid. Studies conducted over the next century brought an abundance of knowledge about viruses, defining them by their inability to replicate outside a host cell, their lack of a protein translation machinery, and their tiny, tiny size, several times smaller than the smallest living bacteria.
Almost 100 years after Ivanovsky’s original finding, a chance observation cast this definition into question. In 1992, scientists investigating a pneumonia outbreak in Bradford, UK, found a strange particle inside an amoeba isolated from water at a cooling plant. At the time, the investigators thought that the particle was a type of gram-positive bacteria, even naming it Bradfordcoccus—for a while. However, attempts to grow it in the laboratory or to amplify its DNA using typical bacterial ribosomal DNA primers ended in failure.
Ten years later, the frozen sample found its way to Marseilles, France. “We tried again to characterize this microbe and we failed,” said Jean-Michel Claverie from the Aix-Marseille University in France. “But we were smart enough—lucky enough—not to throw away the sample, but to look at it under the microscope.”
Electron microscopy proved that against all expectations, this particle was not a bacterium, but a virus. Breaking with the convention of naming viruses after their host, the researchers named this unusually large virus “MIcrobe MImicking Virus”—Mimivirus (1). This discovery opened Pandora’s box, leading researchers to question the very nature of viruses and their place within the tree of life.
Secrets in The Ice
Initially, it appeared as if the 400 nm long Mimivirus may be an anomaly, a biological curiosity with no relatives or parallels. But the closely spaced discoveries of several other giant viruses quickly dispelled this notion. Mimivirus retained its throne as the largest known virus only for a few years, before the discovery of the massive, amphora-shaped Pandoraviruses. Later, Pithovirus sibericum eclipsed Pandoravirus’s place on that list, although Mimivirus still holds the distinction of having one of the largest genomes among giant viruses.
To date, approximately a dozen different strains of mimiviridae and about three strains of pandoroviridiae have been identified in samples collected from all over the world, primarily from natural or man-made water sources.
Perhaps the most fascinating sample yet to yield evidence of giant viruses is a bit of permafrost collected in Siberia that is over 30,000 years old. From this sample, scientists isolated two entirely new kinds of giant viruses that remained infective after being thawed (2,3), raising important questions about future threats to human health from global warming.
The first of the new viruses, named Pithovirus, was a massive 1.5 microns in length. It bears a superficial resemblance to Pandoraviruses, but carries a much smaller genome. The second, named Mollivirus, was distinct from the other three classes of giant viruses in terms of both morphology and genome structure.
Why have these mega-viruses started cropping up all of a sudden? They may have been observed before, but due to their size, were misclassified as bacteria or other microbes, according to Claverie. “There are four families now of giant viruses that have been discovered. At least three of them were spotted by people (when we look at the literature) much before we truly interpreted them as viruses,” said Claverie, who along with his wife Chantal Abergel, was an integral part of the team that discovered the Siberian giant viruses.
The Genome of a Giant
Over the last ten years, research into the biology of giant viruses has progressed rapidly, yielding a basic idea of the replication cycles of all four major classes of giant viruses. The Mimivirus genome sequence was published in 2004 (4). At 1.2 million base pairs, and 911 suspected protein coding genes, the size of the Mimivirus genome was unprecedented amongst viruses.
An in depth analysis of the sequence revealed two surprising findings. First, almost three-fourths of the total identified genes bore absolutely no resemblance to any genes seen in any other organism or virus. Second, the Mimivirus genome had a bunch of genes similar to those used for carrying out protein translation in bacteria and eukaryotes—including seven different amino-acyl tRNA synthetases for linking amino acids to their appropriate tRNA molecules during translation. This was particularly surprising, because just like all other viruses, none of the giant viruses found so far have the ability to carry out translation without a host cell.
The first Pandoravirus sequence was published in 2013 (5), while Mollivirus and Pithovirus were sequenced in 2014 and 2015, respectively (2,3). Even between the different groups of giant viruses, there is surprisingly little similarity in the composition or sequence of genes, and correspondingly in their structure, biology, and mode of replication, even though they all infect the same organism—Acanthamoeba.
All of this gives rise to the big question—where did these giant viruses come from? Were they small like other viruses to begin with, but absorbed extra genes from other organisms they came in contact with, and therefore increased in size to accommodate the increase in DNA content? This idea, referred to as the “accretion theory,” is supported by the presence of a few genes in giant virus genomes that appear to be of bacterial or amoeboid origin, likely acquired by the virus via horizontal gene transfer.
The second theory is that these viruses were once even bigger and free-living, but over millions of years of parasitic history, they progressively lost some genome components, becoming more and more dependent on their hosts, until they reached the form that we see today. This would mean that the translation genes present in their genomes now are left-behind relics of a once-functional translation machinery. “Once a parasite, always a parasite,” said Claverie in support of this theory, “And when you’re a parasite, you become increasingly dependent on your host.”
Parasitizing a Parasite
The final nail in the coffin for the old picture of viruses may be the discovery of virophages—viruses that infect other viruses (6). These satellite viruses are relatively tiny in size, and tend to simultaneously infect the host amoeba alongside Mimiviruses, depending on the host protein-making machinery for their own survival. In the process, some of them interfere with and reduce the infectivity of the mimivirus.
Most interestingly, a putative defense system has been found in the Mimiviruses that researchers named the “Mimiviruses virophage resistance element” (MIMIVIRE) (7). Bearing a certain resemblance to the CRISPR-Cas system found in bacteria, MIMIVIRE contains sequences that may help the virus recognize and then cleave foreign DNA, although this view is contested by others, who believe the Mimiviruses’s defense system may be protein-interaction based instead (8).
Claverie expects the discovery of more such viruses over the next few years. And perhaps with them, we will continue to gain new perspective on the origins of viruses and of life on earth.