Barcoding synapses
Mapping the connectome is perhaps the most ambitious undertaking to date for biomedical research. Will a novel method that precisely labels each synapse help?
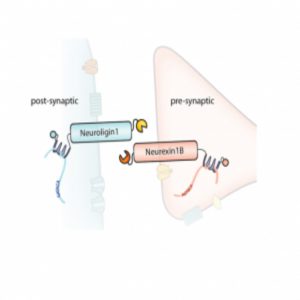
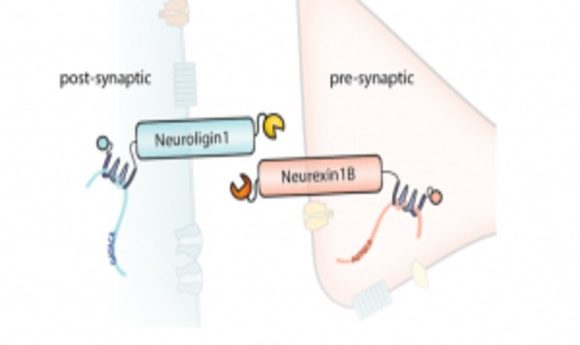
A diagram of the barcoded RNAs, linkers, and pre- and post-synaptic proteins at a synapse.
Credit: Justus Kebschull
Identifying the connections for all the neurons in the brain has perplexed researchers for decades. A solution may soon be at hand, as improved molecular probes and high throughput assays for neurons become more accessible.
A group led by Anthony Zador at Cold Spring Harbor Laboratory has been very active in developing tools that introduce unique random RNA sequences, or barcodes, into neurons in order to map them. Last year, his team reported Multiplexed Analysis of Projections by Sequencing, or MAPseq, where they used deactivated virus to introduce RNA barcodes into a mouse brain to map neuronal projections (1). In a new study in Nucleic Acids Research (2), Zador’s group describes SYNseq, where they incorporated barcodes into pre- and post-synaptic proteins to map synapses, revealing the actual “wiring” of the brain.
“The huge advantage of using sequencing to map synaptic connectivity is that even the currently available sequencing capacity is on par with the complexity of many circuits under study and even entire brains. This is the main reason we are developing SYNseq,” noted Justus Kebschull, lead author of the study.
Kebschull, Zador, and their colleagues used Sindbis virus to express barcodes linked to pre-synaptic or post-synaptic proteins. They also included linker proteins that they could covalently crosslink and fluorescent proteins for visualization. The team confirmed proper expression, membrane localization, and crosslinking in non-neuronal cells, and then used a microfluidic chamber to observe that two distinct primary-cultured neurons with different barcodes could be crosslinked. To sequence the barcodes, they immunoprecipitated the complexes from infected neurons and amplified the target sequences by emulsion PCR; they then used the sequence of each barcode pair to construct a map of the joined neurons.
While Zador’s group was successful in joining pre- and post- synaptic barcodes as a proof of concept, their initial experiments resulted in a high frequency of false positives. Further dilution of the PCR emulsion reduced this; however, they noted that it would also require scaling up SYNseq beyond practical limits for high throughput applications.
Kebschull is optimistic that they will clear these hurdles. “We need better barcode joining and the barcode delivery by viruses might become limiting at some point if dense reconstructions are needed where every cell in the brain needs to be labeled,” he said. “There are viral approaches to overcome this issue, or in vivo generated barcodes could be very powerful in this context.”
While the team designed SYNseq for circuit mapping, “It can be used to map other kinds of cell-to-cell contacts, and could therefore be used to probe the structure of tumors at the single cell level,” Kebschull said.