A solution for true 3D imaging analysis
The pharmaceutical R&D success rate is low, and costs are high, therefore the streamlining of drug discovery processes is highly desired. How can the precision of anti-cancer drug dosage response measurements in a 3D cell-based-assay cancer model be improved?
The research and development (R&D) of pharmaceutical and biotechnology companies worldwide has vigorously developed new drugs to serve unmet medical needs. Generally, 10–20 years and several billion dollars are required to launch a new drug that successfully passes preclinical and clinical tests. As the pharmaceutical R&D success rate is extremely low, reduction in R&D risks and streamlining of drug discovery processes are desired.
Two-dimensional (2D) cell-based assays are often performed before the clinical phases of drug development. The results of the assays do not always reflect those of clinical studies owing to their artificial culture conditions. To overcome this issue, three-dimensional (3D) cell culture models have garnered attention for drug evaluation. 3D cell culture models, such as spheroids or organoids produced by hanging drop methods or low adhesion plates, can mimic key factors of tissues and are more representative of an in vivo microenvironment than 2D monolayer culture models because of the exhibition of cell–cell interactions and gradients of oxygen or nutrients. Utilizing these 3D culture models can contribute to more in vivo-like drug evaluation; therefore, we could obtain more predictive results of compound efficacy before the clinical phase.
3D cell culture models comprise the heterogeneous cellular population and abundant information of in vivo tumors or tissues. When identifying compound efficacy using these 3D cell culture models, highly accurate and quantitative methods are required to evaluate the compounds’ targets. One existing method for quantifying compound efficacy in 3D models is the use of a plate screening luminometer to measure ATP. Changes in ATP levels are thought to correlate with the viability of the 3D models treated with compounds, however this method loses spatial information of the 3D models such as cellular localization or morphological structure. To gain insights into the mode of the compounds’ action, imaging technology and analysis methods are important. Conventional imaging analysis methods for 3D cell culture models involve a wide-field image or a projection image that is produced by overlapping a series of plane images acquired by confocal systems. Hereinafter, we refer to the former as “2D analysis” and the latter as “2.5D analysis.” It is desired to obtain their spatial information, single cell localization, or organelle conditions quantitatively and combine this multiparametric quantitative information for the evaluation of 3D cell culture models such that new insights of compounds can be attained. We term this 3D imaging-analysis workflow as “True 3D analysis” (Figure 1).
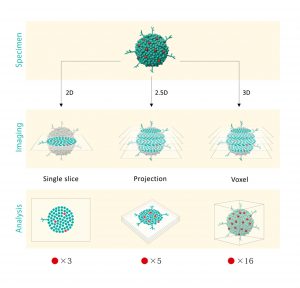
Figure 1. Scheme of 2D, 2.5D, and “True 3D analysis”
We created a theoretical model spheroid that comprises 16 red cell objects of interest. 2D analysis uses a single z-slice image and three red objects can be found in the specimen. 2.5D analysis uses a stacked image and five red objects can be found because the objects along the same Z-axis cannot be separated in a maximum intensity projection. Furthermore, the morphology of the detected objects in 2.5D have been distorted since multiple cell objects contribute to the projected image. Only “True 3D analysis” can recognize 16 red objects that are identical to the original model, with accurate morphology.
We developed a unique 3D high content analysis software, “NoviSight” for “True 3D analysis.” This software enables the rapid performance of the whole structure analysis, cell segmentation, individual organelle recognition, and their quantification across multiple specimens in microplates. Furthermore, the calculated quantitative data can be sorted graphically by multiple parameters. Herein, we introduce the utility of the “True 3D analysis” workflow established by the combination of a confocal imaging system and quantitative 3D analysis in a 3D gel invasion assay as an example.
Tumors spread to remote organs through blood circulation or lymphatic vessels. As tumor metastasis is a major prognostic indicator for patient mortality, it is regarded as an important target for anti-cancer drug development. Metastatic cells exhibit weak adhesion and gain active motility. These highly motile cells enter the vasculature after having successfully crossed the surrounding tissues. Once they enter the blood or lymphatic flow, tumors are arrested with a remote capillary, migrate into extravascular tissues, and finally form new metastatic lesions. Therefore, invasion, the first step in metastasis, is a fundamental step in tumor progression and the driving force for metastasis.
The 3D gel invasion assay has gained popularity in drug discovery research. Typically, a wound-healing assay or trans-well assay is used for the evaluation of tumor invasion. Both of these traditional assays are simple and inexpensive, but the disadvantage of these methods is that they lack a tumor microenvironment, and cells lose their relevant properties, such as differentiation, polarization, and cell–cell communication. Therefore, these useful but artificial assays contribute to the poor predictive value of compound efficacies between in vitro and in vivo experiments.
A 3D gel tumor invasion assay can recreate the tumor microenvironment well by embedding tumor spheroids into a gel matrix. The cells in the embedded spheroids sprout gradually into the gel. This assay can mimic tumor morphology, collectiveness of invasion, or epithelial–mesenchymal transition following invasion activity such that using this system renders a more in vivo-like drug evaluation.
We used this 3D gel tumor invasion assay to form human fibrosarcoma cell HT1080 spheroids in a 96well U plate and embedded them into a Matrigel matrix (CorningR). To test the drug efficacy against invasion, we used a strong protein kinase C (PKC) inhibitor Staurosporin (STS) or matrix metalloprotease-specific inhibitor Batimastat (BB94). The drug efficacies were evaluated by the “True 3D” quantification of cell viability and morphological changes.
First, to visualize the HT1080 spheroids’ morphology, we stained the HT1080 cells’ actin with Acti-stainTM 670(Cytoskelton, Inc.) and performed imaging using a confocal laser scanning microscope FV3000. We found that nontreated HT1080 cells invaded the matrigel three dimensionally, while STS and BB94 inhibited HT1080 invasion (Fig. 2).
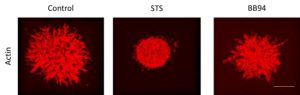
Figure 2. Drug efficacy in HT1080 invasion
HT1080 invasive spheroids were stained with Actistain670. Scale: 100 µm
The HT1080 invasive spheroids were structurally divided into two components: invasive volume and spheroid core volume, and NoviSight could recognize them respectively. Only “True 3D analysis” could quantify the whole morphology including invasion in the z-axis volume which would be lost in 2D or 2.5D analysis. When we performed the recognition of their respective invasive volume and core volume (Fig. 3A), and quantified their morphologies by NoviSight, we found that STS and BB94 decreased HT1080 invasion in a dose-dependent manner (Fig. 3B).
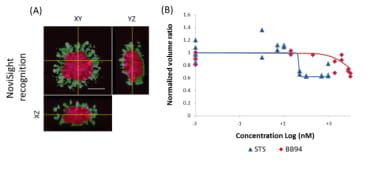
Figure 3. Volume recognition and quantification by “True 3D analysis”
(A) NoviSight can recognize and quantify the invasive volume (green area) and spheroid core volume (red), separately. Scale: 100 µm (B) Dose-response plot of STS and BB94 for HT1080 invasion volume quantified by NoviSight. We normalized the invasion-to-core volume ratio at each concentration with the control.
Subsequently, we stained the HT1080 spheroids with the NUCLEAR-IDR Red/Green cell viability reagent (Enzo) and imaged the Z-series of the samples by FV3000 to perform a cell viability test. This reagent stains the live cells’ nuclei with the red and dead cells’ nuclei with green. BB94-treated HT1080 cells and the nontreated cells were almost alive, while STS induced HT1080 cell apoptosis (Fig. 4).

Figure 4. 3D volume rendering of HT1080 invasion in viability test
2μM STS caused HT1080 cell apoptosis while 20μM BB94 did not affect HT1080 cell viability. Scale: 100 µm
A general 2.5D analysis was performed based on the superposed images; however, the pixels located in the same z-axis were compressed such that the 2.5D analysis could not yield a single-cell recognition. In contrast, NoviSight’s “True 3D analysis” can recognize spatial nucleus localization(Fig. 5A). By using this characteristic function of NoviSight, we counted the number of live/dead stained cells and found that STS induced HT1080 apoptosis at above 50 nM, and BB94 did not influence HT1080 viability until 20 µM (Fig. 5B). In general, a spheroid viability test is performed using the CellTiter-GloR3D Cell Viability Assay (Promega) and the lysates are measured using a luminometer. We also tested this assay to compare with the viability results of “True 3D analysis” and found that the compounds’ specificity analyzed by the NoviSight and Glo assays were almost the same; STS affected cell viability but BB94 did not (Fig. 5c). The viability curves were different between the NoviSight and CellTiter-Glo assays. This difference may be caused by the properties measured using the analysis methods; NoviSight quantified the cell viability for each cell, while the CellTiter-Glo assay quantified the total ATP amounts of the spheroids.
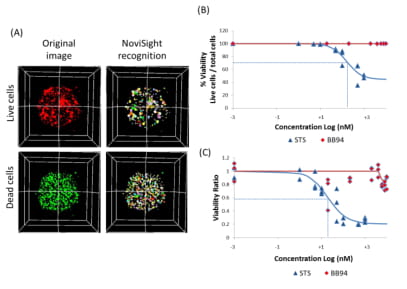 Figure 5. Nuclear recognition and quantification by “True 3D analysis”
Figure 5. Nuclear recognition and quantification by “True 3D analysis”
(A) NoviSight can perform live or dead cells’ nuclear recognition and counting. Each pseudo-colored object is a HT1080 cell nucleus. (B, C) Dose-response plot of STS and BB94 for HT1080 cell viability by NoviSight (B) and CellTiter-Glo assay (C). IC50 values for STS were 160.3nM by NoviSight while 19.3nM by CellTiter-Glo assay.
We performed a drug efficacy study for 3D gel invasion models using “True 3D analysis” and achieved the evaluation of HT1080 cell viability coupled with morphological changes. Only imaging-based cell analysis enabled us to estimate drug efficacy with multiple parameters which included structural information and nuclei object counts. In this study, even the effective concentration of BB94 for HT1080 invasion did not affect HT1080 cell death unlike the case of STS. These data can contribute to the accurate prediction of the compounds’ specificity.
The novel “True 3D analysis,” and the combination with the acquisition of high-resolution images and 3D analysis, enabled us to perform a single-cell level analysis of the 3D models while measuring their morphologies. We could capture the cellular localization, morphology, or heterogeneity of the complex 3D models that conventional assays might not. There is a provocative difference in measured dose dependency detected using NoviSight and True 3D methods as compared to non-imaging methods. Actual single cell and nuclear measurements are enabled by full 3D analysis. Therefore, we believe that “True 3D analysis” can contribute to a precise drug evaluation performed using cell-based assays.